
Pectinobacterium phage PEAT2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peatvirus; Pectinobacterium virus PEAT2
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
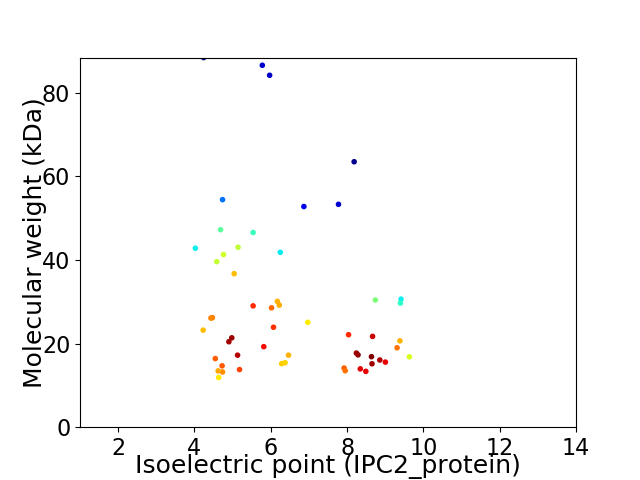
Virtual 2D-PAGE plot for 55 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
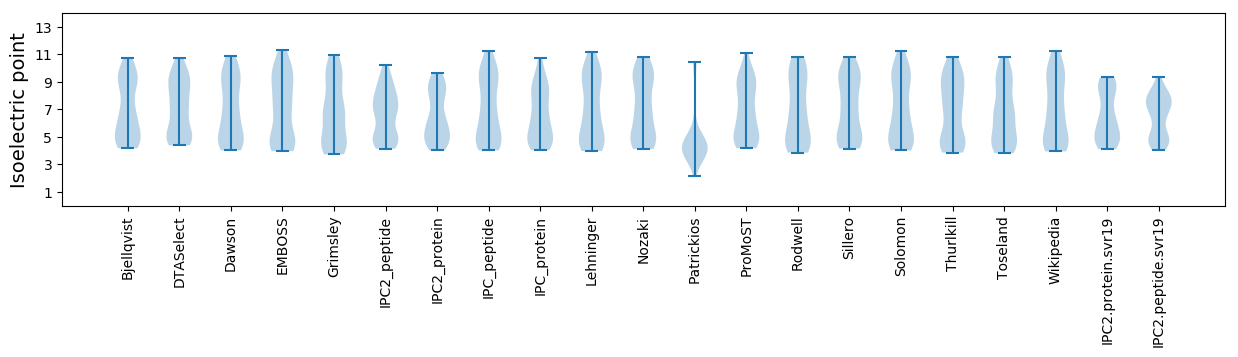
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4N7B0|A0A2H4N7B0_9CAUD Tail fiber protein OS=Pectinobacterium phage PEAT2 OX=2053078 PE=4 SV=1
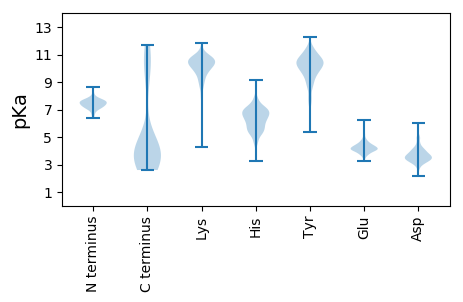
MM1 pKa = 7.92AEE3 pKa = 4.19LTSTGYY9 pKa = 9.8SVKK12 pKa = 10.61SQNDD16 pKa = 3.18WFDD19 pKa = 3.78EE20 pKa = 4.22EE21 pKa = 4.11QQLYY25 pKa = 10.95RR26 pKa = 11.84NIDD29 pKa = 3.89SNWNLDD35 pKa = 3.15PSTPDD40 pKa = 3.47GLKK43 pKa = 9.09MAHH46 pKa = 6.79DD47 pKa = 4.33AEE49 pKa = 4.65IFSALDD55 pKa = 3.24EE56 pKa = 4.48VLQQAYY62 pKa = 10.17NSKK65 pKa = 10.83DD66 pKa = 3.36PNKK69 pKa = 10.71ASGYY73 pKa = 10.2DD74 pKa = 3.51LDD76 pKa = 4.94VICALTGTVRR86 pKa = 11.84SEE88 pKa = 4.03GTASTVTGFVLTGTPGTQVPAGTRR112 pKa = 11.84FEE114 pKa = 4.52SSVTGYY120 pKa = 10.68RR121 pKa = 11.84FALDD125 pKa = 3.44QTWTLDD131 pKa = 3.37SSGTATVDD139 pKa = 3.23ITCTTVGEE147 pKa = 4.34IEE149 pKa = 4.58ADD151 pKa = 3.23ANTITTIVDD160 pKa = 3.73TVAGLVSVNNPTPATPGTAAEE181 pKa = 4.37SDD183 pKa = 3.2GSLRR187 pKa = 11.84LKK189 pKa = 10.49RR190 pKa = 11.84ATAVGLPGSNQVDD203 pKa = 3.73SMLGQLFNVDD213 pKa = 2.55GVRR216 pKa = 11.84RR217 pKa = 11.84VRR219 pKa = 11.84VYY221 pKa = 11.13EE222 pKa = 4.0NDD224 pKa = 3.23EE225 pKa = 4.11AATDD229 pKa = 3.92SNGQPGHH236 pKa = 6.57SIAPIIDD243 pKa = 3.7GGTDD247 pKa = 3.56DD248 pKa = 4.92DD249 pKa = 4.12VAMAIYY255 pKa = 10.12LKK257 pKa = 10.46KK258 pKa = 10.73NPGVTLYY265 pKa = 10.68QAGTDD270 pKa = 3.41VTVTVTSPTYY280 pKa = 8.95PTMTKK285 pKa = 9.97DD286 pKa = 3.14IKK288 pKa = 10.47FSRR291 pKa = 11.84PVYY294 pKa = 10.01VDD296 pKa = 2.79MVVAIEE302 pKa = 4.23IKK304 pKa = 10.56DD305 pKa = 3.92DD306 pKa = 3.79GSLPSQVTLEE316 pKa = 4.45PLIQDD321 pKa = 4.96AIMEE325 pKa = 4.3YY326 pKa = 10.66AAGGLIPTEE335 pKa = 4.23YY336 pKa = 10.73GFKK339 pKa = 10.12PDD341 pKa = 3.92GFDD344 pKa = 2.96IGEE347 pKa = 4.3TVPYY351 pKa = 10.09SSLYY355 pKa = 9.26TPINKK360 pKa = 9.51VIGSYY365 pKa = 10.9GNSYY369 pKa = 9.48VNSMTLNSGTANVTIDD385 pKa = 3.64FNEE388 pKa = 4.07LSRR391 pKa = 11.84WTASNITVTIVV402 pKa = 2.6
MM1 pKa = 7.92AEE3 pKa = 4.19LTSTGYY9 pKa = 9.8SVKK12 pKa = 10.61SQNDD16 pKa = 3.18WFDD19 pKa = 3.78EE20 pKa = 4.22EE21 pKa = 4.11QQLYY25 pKa = 10.95RR26 pKa = 11.84NIDD29 pKa = 3.89SNWNLDD35 pKa = 3.15PSTPDD40 pKa = 3.47GLKK43 pKa = 9.09MAHH46 pKa = 6.79DD47 pKa = 4.33AEE49 pKa = 4.65IFSALDD55 pKa = 3.24EE56 pKa = 4.48VLQQAYY62 pKa = 10.17NSKK65 pKa = 10.83DD66 pKa = 3.36PNKK69 pKa = 10.71ASGYY73 pKa = 10.2DD74 pKa = 3.51LDD76 pKa = 4.94VICALTGTVRR86 pKa = 11.84SEE88 pKa = 4.03GTASTVTGFVLTGTPGTQVPAGTRR112 pKa = 11.84FEE114 pKa = 4.52SSVTGYY120 pKa = 10.68RR121 pKa = 11.84FALDD125 pKa = 3.44QTWTLDD131 pKa = 3.37SSGTATVDD139 pKa = 3.23ITCTTVGEE147 pKa = 4.34IEE149 pKa = 4.58ADD151 pKa = 3.23ANTITTIVDD160 pKa = 3.73TVAGLVSVNNPTPATPGTAAEE181 pKa = 4.37SDD183 pKa = 3.2GSLRR187 pKa = 11.84LKK189 pKa = 10.49RR190 pKa = 11.84ATAVGLPGSNQVDD203 pKa = 3.73SMLGQLFNVDD213 pKa = 2.55GVRR216 pKa = 11.84RR217 pKa = 11.84VRR219 pKa = 11.84VYY221 pKa = 11.13EE222 pKa = 4.0NDD224 pKa = 3.23EE225 pKa = 4.11AATDD229 pKa = 3.92SNGQPGHH236 pKa = 6.57SIAPIIDD243 pKa = 3.7GGTDD247 pKa = 3.56DD248 pKa = 4.92DD249 pKa = 4.12VAMAIYY255 pKa = 10.12LKK257 pKa = 10.46KK258 pKa = 10.73NPGVTLYY265 pKa = 10.68QAGTDD270 pKa = 3.41VTVTVTSPTYY280 pKa = 8.95PTMTKK285 pKa = 9.97DD286 pKa = 3.14IKK288 pKa = 10.47FSRR291 pKa = 11.84PVYY294 pKa = 10.01VDD296 pKa = 2.79MVVAIEE302 pKa = 4.23IKK304 pKa = 10.56DD305 pKa = 3.92DD306 pKa = 3.79GSLPSQVTLEE316 pKa = 4.45PLIQDD321 pKa = 4.96AIMEE325 pKa = 4.3YY326 pKa = 10.66AAGGLIPTEE335 pKa = 4.23YY336 pKa = 10.73GFKK339 pKa = 10.12PDD341 pKa = 3.92GFDD344 pKa = 2.96IGEE347 pKa = 4.3TVPYY351 pKa = 10.09SSLYY355 pKa = 9.26TPINKK360 pKa = 9.51VIGSYY365 pKa = 10.9GNSYY369 pKa = 9.48VNSMTLNSGTANVTIDD385 pKa = 3.64FNEE388 pKa = 4.07LSRR391 pKa = 11.84WTASNITVTIVV402 pKa = 2.6
Molecular weight: 42.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4N7D0|A0A2H4N7D0_9CAUD Uncharacterized protein OS=Pectinobacterium phage PEAT2 OX=2053078 PE=4 SV=1
MM1 pKa = 7.56SFGLRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84HH10 pKa = 4.89VPSAFSALLVEE21 pKa = 5.54WIRR24 pKa = 11.84SSHH27 pKa = 5.05GVPVEE32 pKa = 4.25STGSAISSTVAMYY45 pKa = 9.55PCEE48 pKa = 4.23KK49 pKa = 10.32LPSPTGSGSPTNFCSVIMCVAFGTTSASTCCAVVSISGSLRR90 pKa = 11.84LPCNSSPSKK99 pKa = 10.28RR100 pKa = 11.84ASCIMRR106 pKa = 11.84GAAFINSPVAVFKK119 pKa = 10.89RR120 pKa = 11.84SNAAVASVVSMFNLYY135 pKa = 10.37DD136 pKa = 3.48SSPAVVLRR144 pKa = 11.84GSAPCASAISSSALPNARR162 pKa = 11.84SAMWSSFGMVKK173 pKa = 10.21VIIIWTPVVRR183 pKa = 11.84ILHH186 pKa = 5.68ACKK189 pKa = 9.99KK190 pKa = 10.1LRR192 pKa = 11.84YY193 pKa = 8.98LRR195 pKa = 11.84CC196 pKa = 3.75
MM1 pKa = 7.56SFGLRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84HH10 pKa = 4.89VPSAFSALLVEE21 pKa = 5.54WIRR24 pKa = 11.84SSHH27 pKa = 5.05GVPVEE32 pKa = 4.25STGSAISSTVAMYY45 pKa = 9.55PCEE48 pKa = 4.23KK49 pKa = 10.32LPSPTGSGSPTNFCSVIMCVAFGTTSASTCCAVVSISGSLRR90 pKa = 11.84LPCNSSPSKK99 pKa = 10.28RR100 pKa = 11.84ASCIMRR106 pKa = 11.84GAAFINSPVAVFKK119 pKa = 10.89RR120 pKa = 11.84SNAAVASVVSMFNLYY135 pKa = 10.37DD136 pKa = 3.48SSPAVVLRR144 pKa = 11.84GSAPCASAISSSALPNARR162 pKa = 11.84SAMWSSFGMVKK173 pKa = 10.21VIIIWTPVVRR183 pKa = 11.84ILHH186 pKa = 5.68ACKK189 pKa = 9.99KK190 pKa = 10.1LRR192 pKa = 11.84YY193 pKa = 8.98LRR195 pKa = 11.84CC196 pKa = 3.75
Molecular weight: 20.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14633 |
107 |
820 |
266.1 |
29.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.939 ± 0.38 | 1.633 ± 0.199 |
6.109 ± 0.249 | 5.078 ± 0.281 |
3.724 ± 0.202 | 7.456 ± 0.37 |
1.989 ± 0.217 | 5.74 ± 0.191 |
4.77 ± 0.332 | 7.469 ± 0.247 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.857 ± 0.17 | 4.852 ± 0.279 |
4.353 ± 0.228 | 3.82 ± 0.258 |
5.597 ± 0.335 | 6.465 ± 0.309 |
6.745 ± 0.304 | 7.497 ± 0.262 |
1.667 ± 0.141 | 3.239 ± 0.146 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
