
BtMf-AlphaCoV/GD2012
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Minunacovirus; Miniopterus bat coronavirus HKU8
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
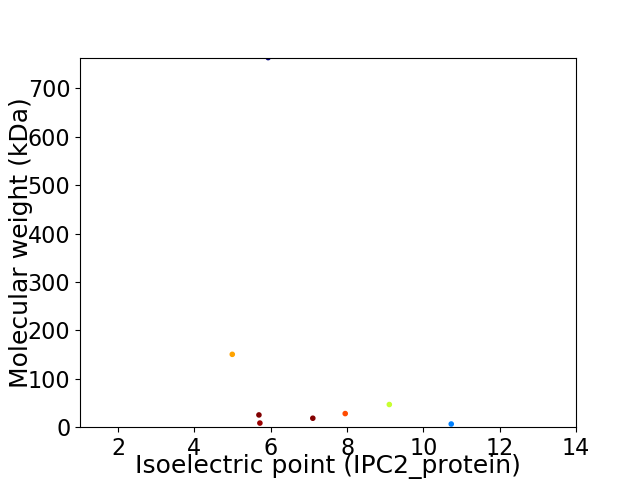
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U1WHC0|A0A0U1WHC0_9ALPC Uncharacterized protein OS=BtMf-AlphaCoV/GD2012 OX=1503280 PE=4 SV=1
MM1 pKa = 7.68LFLFLVNVAFAANQDD16 pKa = 3.66CTGNGNLDD24 pKa = 3.41SMEE27 pKa = 4.41DD28 pKa = 3.53LKK30 pKa = 11.71LNLPPNVTNAYY41 pKa = 9.54VSGYY45 pKa = 10.63LPLPQQWNCSVSGTLGNPRR64 pKa = 11.84VYY66 pKa = 10.19TGAHH70 pKa = 5.33GVFVSYY76 pKa = 10.88YY77 pKa = 10.82DD78 pKa = 3.33SATDD82 pKa = 3.25IVVGVGSTLSNSSWGMYY99 pKa = 9.2FFQKK103 pKa = 10.32NSEE106 pKa = 3.48GRR108 pKa = 11.84AIVRR112 pKa = 11.84ICKK115 pKa = 6.99WTSFNTPNYY124 pKa = 8.57QPNVNTQGRR133 pKa = 11.84DD134 pKa = 3.4CLFNKK139 pKa = 9.57QFSFTFIHH147 pKa = 6.42NGNEE151 pKa = 3.69IVGASWAGDD160 pKa = 3.28KK161 pKa = 10.87VVLYY165 pKa = 8.07TRR167 pKa = 11.84RR168 pKa = 11.84GLTTFYY174 pKa = 11.33VPGAEE179 pKa = 4.03FWDD182 pKa = 4.19VVSIRR187 pKa = 11.84CKK189 pKa = 10.81NKK191 pKa = 9.59GACAHH196 pKa = 5.59QVITSQSTVIVSTNGRR212 pKa = 11.84GLISNYY218 pKa = 7.15TLCSNCSGFPQHH230 pKa = 6.0VFAVGPDD237 pKa = 3.63GSIPPNFEE245 pKa = 3.62FEE247 pKa = 3.78NWFYY251 pKa = 10.55LTNSSSPVSGKK262 pKa = 9.85FVSVQPLQLACLWPIPALLGNTLPVYY288 pKa = 10.92FNMSNNNANCNGFKK302 pKa = 10.56ADD304 pKa = 3.76GVVDD308 pKa = 3.76SLRR311 pKa = 11.84FALNFSDD318 pKa = 3.24EE319 pKa = 4.13RR320 pKa = 11.84AFARR324 pKa = 11.84EE325 pKa = 4.09GVISLEE331 pKa = 4.52TVGNTYY337 pKa = 11.14NFTCTNSSTPTDD349 pKa = 3.44GSSFVLPFGVIDD361 pKa = 3.42QAYY364 pKa = 7.73YY365 pKa = 11.32CFMTFYY371 pKa = 11.26LNRR374 pKa = 11.84TADD377 pKa = 3.37LKK379 pKa = 8.28TTVFAGMLPPVVRR392 pKa = 11.84EE393 pKa = 3.72FVVMRR398 pKa = 11.84NGDD401 pKa = 3.5FYY403 pKa = 11.84LNGFRR408 pKa = 11.84VFSVGQVVSARR419 pKa = 11.84FNISSNDD426 pKa = 3.22SRR428 pKa = 11.84DD429 pKa = 3.15FWTVAFANNVPVLVEE444 pKa = 4.0INSTNIQQLLYY455 pKa = 10.73CNSPLNTIKK464 pKa = 10.65CQQLRR469 pKa = 11.84FNLDD473 pKa = 3.01NGFYY477 pKa = 10.35SYY479 pKa = 11.18SPQVNHH485 pKa = 6.27EE486 pKa = 4.18LPRR489 pKa = 11.84TVVRR493 pKa = 11.84LPKK496 pKa = 10.52FMTHH500 pKa = 5.09SVVNVSVAVSYY511 pKa = 11.16DD512 pKa = 3.34VVNQTNLGLIAWRR525 pKa = 11.84LSFADD530 pKa = 4.02HH531 pKa = 6.51WYY533 pKa = 10.48GWASDD538 pKa = 3.5NRR540 pKa = 11.84TTVCVDD546 pKa = 3.49TSSFTVKK553 pKa = 10.51LFTGSFSYY561 pKa = 11.09GSVFVDD567 pKa = 3.77VLAGEE572 pKa = 4.81CPFTLLSLNDD582 pKa = 3.63YY583 pKa = 11.03LSFGSLCLSLVPNGGCAMSVVTRR606 pKa = 11.84GSYY609 pKa = 10.88GEE611 pKa = 4.08PQSIAPLYY619 pKa = 10.81VSFSDD624 pKa = 3.46GDD626 pKa = 3.81NVIGVPKK633 pKa = 9.8PPPPGVLDD641 pKa = 3.75MSDD644 pKa = 3.04VYY646 pKa = 11.87YY647 pKa = 10.61NVCTSYY653 pKa = 10.57TIYY656 pKa = 10.56GHH658 pKa = 5.65TGRR661 pKa = 11.84GVISKK666 pKa = 10.13GAVDD670 pKa = 4.41FVSGLFYY677 pKa = 10.61TSPAGDD683 pKa = 3.45LLAYY687 pKa = 9.79KK688 pKa = 10.47NSTTGEE694 pKa = 3.91VFMIQPCQFASQVAVIADD712 pKa = 4.6TIVGAASSSANSSIPFNNTVDD733 pKa = 3.47AGLFYY738 pKa = 10.96YY739 pKa = 10.54HH740 pKa = 7.0FNNVTDD746 pKa = 4.05VNGTKK751 pKa = 10.53CEE753 pKa = 4.15VPTLTYY759 pKa = 10.66GGLGICADD767 pKa = 3.9GKK769 pKa = 10.41IVNATRR775 pKa = 11.84RR776 pKa = 11.84YY777 pKa = 8.69AAPEE781 pKa = 3.83PVSPVITGNISVPMNFSFSVQVEE804 pKa = 4.35YY805 pKa = 10.52IQIMLKK811 pKa = 10.27PVTVDD816 pKa = 2.84CSVYY820 pKa = 10.32VCNGNPRR827 pKa = 11.84CLQLLSQYY835 pKa = 11.5ASACKK840 pKa = 9.77TIEE843 pKa = 3.88QALQLSARR851 pKa = 11.84LEE853 pKa = 4.32SVEE856 pKa = 4.18VNSMVAISDD865 pKa = 3.53TALNLGVISKK875 pKa = 10.46FDD877 pKa = 3.37NTFNLSNVLPASVGGKK893 pKa = 8.93SVIEE897 pKa = 4.3DD898 pKa = 3.72LLFDD902 pKa = 3.94KK903 pKa = 11.1VVTSGLGTVDD913 pKa = 3.3TDD915 pKa = 3.77YY916 pKa = 11.21KK917 pKa = 11.24ACAEE921 pKa = 4.05KK922 pKa = 9.97MANTIAEE929 pKa = 4.49AGCVQYY935 pKa = 11.5YY936 pKa = 9.78NGIMVLPGVVDD947 pKa = 3.64QSLLGQYY954 pKa = 7.73TAALTGAMVMGGITAAAAIPFSIAVQSRR982 pKa = 11.84LNYY985 pKa = 10.06LALQTDD991 pKa = 4.07VLQRR995 pKa = 11.84NQQILAQSFNSAMGNITNAFEE1016 pKa = 4.4NVGNAIEE1023 pKa = 4.12QTSQGLQTVAQALDD1037 pKa = 3.47KK1038 pKa = 11.0VQNAVNEE1045 pKa = 4.12QGNALSHH1052 pKa = 5.84LTKK1055 pKa = 10.55QLASNFQAISASIEE1069 pKa = 3.88DD1070 pKa = 4.01LYY1072 pKa = 11.8VRR1074 pKa = 11.84LGQVEE1079 pKa = 4.59ADD1081 pKa = 3.54QQVDD1085 pKa = 3.49RR1086 pKa = 11.84LITGRR1091 pKa = 11.84LAALNAFVSQQLTKK1105 pKa = 9.57YY1106 pKa = 8.06TDD1108 pKa = 3.16VRR1110 pKa = 11.84ASRR1113 pKa = 11.84QLAQEE1118 pKa = 4.75KK1119 pKa = 10.45INEE1122 pKa = 4.44CVKK1125 pKa = 10.63SQSFRR1130 pKa = 11.84YY1131 pKa = 8.66GFCGNGTHH1139 pKa = 5.87VFSVANAAPDD1149 pKa = 4.35GIMFFHH1155 pKa = 7.06SVLLPTAYY1163 pKa = 10.48AEE1165 pKa = 4.25VNAYY1169 pKa = 10.26AGLCVDD1175 pKa = 3.32YY1176 pKa = 10.6KK1177 pKa = 11.25GYY1179 pKa = 10.42VLRR1182 pKa = 11.84DD1183 pKa = 3.44PGNVLFQKK1191 pKa = 10.27PGNEE1195 pKa = 4.26TYY1197 pKa = 10.87LITPRR1202 pKa = 11.84RR1203 pKa = 11.84MFEE1206 pKa = 3.66PRR1208 pKa = 11.84VPQMSDD1214 pKa = 2.99FVQISGCNVSYY1225 pKa = 11.53VNVSSEE1231 pKa = 4.04QLPDD1235 pKa = 3.41IVPDD1239 pKa = 4.35FMDD1242 pKa = 3.64VNATIQDD1249 pKa = 3.82ILDD1252 pKa = 3.66KK1253 pKa = 10.5LQNYY1257 pKa = 7.61SKK1259 pKa = 10.75PDD1261 pKa = 3.56FGLDD1265 pKa = 3.12IFNATYY1271 pKa = 11.05LNLSAEE1277 pKa = 4.48INDD1280 pKa = 3.59LTNRR1284 pKa = 11.84SQQLQAITDD1293 pKa = 3.75EE1294 pKa = 4.24LRR1296 pKa = 11.84ATIANINGTLVDD1308 pKa = 4.67LEE1310 pKa = 4.09WLNRR1314 pKa = 11.84VEE1316 pKa = 5.97TYY1318 pKa = 10.4IKK1320 pKa = 9.3WPWYY1324 pKa = 7.96VWLAIAVALIVLIGLMLWCCLSTGCCGCCSCMASTLDD1361 pKa = 3.63FRR1363 pKa = 11.84GSRR1366 pKa = 11.84LQQYY1370 pKa = 7.6EE1371 pKa = 4.15VEE1373 pKa = 4.46KK1374 pKa = 11.13VHH1376 pKa = 6.56IQQ1378 pKa = 3.09
MM1 pKa = 7.68LFLFLVNVAFAANQDD16 pKa = 3.66CTGNGNLDD24 pKa = 3.41SMEE27 pKa = 4.41DD28 pKa = 3.53LKK30 pKa = 11.71LNLPPNVTNAYY41 pKa = 9.54VSGYY45 pKa = 10.63LPLPQQWNCSVSGTLGNPRR64 pKa = 11.84VYY66 pKa = 10.19TGAHH70 pKa = 5.33GVFVSYY76 pKa = 10.88YY77 pKa = 10.82DD78 pKa = 3.33SATDD82 pKa = 3.25IVVGVGSTLSNSSWGMYY99 pKa = 9.2FFQKK103 pKa = 10.32NSEE106 pKa = 3.48GRR108 pKa = 11.84AIVRR112 pKa = 11.84ICKK115 pKa = 6.99WTSFNTPNYY124 pKa = 8.57QPNVNTQGRR133 pKa = 11.84DD134 pKa = 3.4CLFNKK139 pKa = 9.57QFSFTFIHH147 pKa = 6.42NGNEE151 pKa = 3.69IVGASWAGDD160 pKa = 3.28KK161 pKa = 10.87VVLYY165 pKa = 8.07TRR167 pKa = 11.84RR168 pKa = 11.84GLTTFYY174 pKa = 11.33VPGAEE179 pKa = 4.03FWDD182 pKa = 4.19VVSIRR187 pKa = 11.84CKK189 pKa = 10.81NKK191 pKa = 9.59GACAHH196 pKa = 5.59QVITSQSTVIVSTNGRR212 pKa = 11.84GLISNYY218 pKa = 7.15TLCSNCSGFPQHH230 pKa = 6.0VFAVGPDD237 pKa = 3.63GSIPPNFEE245 pKa = 3.62FEE247 pKa = 3.78NWFYY251 pKa = 10.55LTNSSSPVSGKK262 pKa = 9.85FVSVQPLQLACLWPIPALLGNTLPVYY288 pKa = 10.92FNMSNNNANCNGFKK302 pKa = 10.56ADD304 pKa = 3.76GVVDD308 pKa = 3.76SLRR311 pKa = 11.84FALNFSDD318 pKa = 3.24EE319 pKa = 4.13RR320 pKa = 11.84AFARR324 pKa = 11.84EE325 pKa = 4.09GVISLEE331 pKa = 4.52TVGNTYY337 pKa = 11.14NFTCTNSSTPTDD349 pKa = 3.44GSSFVLPFGVIDD361 pKa = 3.42QAYY364 pKa = 7.73YY365 pKa = 11.32CFMTFYY371 pKa = 11.26LNRR374 pKa = 11.84TADD377 pKa = 3.37LKK379 pKa = 8.28TTVFAGMLPPVVRR392 pKa = 11.84EE393 pKa = 3.72FVVMRR398 pKa = 11.84NGDD401 pKa = 3.5FYY403 pKa = 11.84LNGFRR408 pKa = 11.84VFSVGQVVSARR419 pKa = 11.84FNISSNDD426 pKa = 3.22SRR428 pKa = 11.84DD429 pKa = 3.15FWTVAFANNVPVLVEE444 pKa = 4.0INSTNIQQLLYY455 pKa = 10.73CNSPLNTIKK464 pKa = 10.65CQQLRR469 pKa = 11.84FNLDD473 pKa = 3.01NGFYY477 pKa = 10.35SYY479 pKa = 11.18SPQVNHH485 pKa = 6.27EE486 pKa = 4.18LPRR489 pKa = 11.84TVVRR493 pKa = 11.84LPKK496 pKa = 10.52FMTHH500 pKa = 5.09SVVNVSVAVSYY511 pKa = 11.16DD512 pKa = 3.34VVNQTNLGLIAWRR525 pKa = 11.84LSFADD530 pKa = 4.02HH531 pKa = 6.51WYY533 pKa = 10.48GWASDD538 pKa = 3.5NRR540 pKa = 11.84TTVCVDD546 pKa = 3.49TSSFTVKK553 pKa = 10.51LFTGSFSYY561 pKa = 11.09GSVFVDD567 pKa = 3.77VLAGEE572 pKa = 4.81CPFTLLSLNDD582 pKa = 3.63YY583 pKa = 11.03LSFGSLCLSLVPNGGCAMSVVTRR606 pKa = 11.84GSYY609 pKa = 10.88GEE611 pKa = 4.08PQSIAPLYY619 pKa = 10.81VSFSDD624 pKa = 3.46GDD626 pKa = 3.81NVIGVPKK633 pKa = 9.8PPPPGVLDD641 pKa = 3.75MSDD644 pKa = 3.04VYY646 pKa = 11.87YY647 pKa = 10.61NVCTSYY653 pKa = 10.57TIYY656 pKa = 10.56GHH658 pKa = 5.65TGRR661 pKa = 11.84GVISKK666 pKa = 10.13GAVDD670 pKa = 4.41FVSGLFYY677 pKa = 10.61TSPAGDD683 pKa = 3.45LLAYY687 pKa = 9.79KK688 pKa = 10.47NSTTGEE694 pKa = 3.91VFMIQPCQFASQVAVIADD712 pKa = 4.6TIVGAASSSANSSIPFNNTVDD733 pKa = 3.47AGLFYY738 pKa = 10.96YY739 pKa = 10.54HH740 pKa = 7.0FNNVTDD746 pKa = 4.05VNGTKK751 pKa = 10.53CEE753 pKa = 4.15VPTLTYY759 pKa = 10.66GGLGICADD767 pKa = 3.9GKK769 pKa = 10.41IVNATRR775 pKa = 11.84RR776 pKa = 11.84YY777 pKa = 8.69AAPEE781 pKa = 3.83PVSPVITGNISVPMNFSFSVQVEE804 pKa = 4.35YY805 pKa = 10.52IQIMLKK811 pKa = 10.27PVTVDD816 pKa = 2.84CSVYY820 pKa = 10.32VCNGNPRR827 pKa = 11.84CLQLLSQYY835 pKa = 11.5ASACKK840 pKa = 9.77TIEE843 pKa = 3.88QALQLSARR851 pKa = 11.84LEE853 pKa = 4.32SVEE856 pKa = 4.18VNSMVAISDD865 pKa = 3.53TALNLGVISKK875 pKa = 10.46FDD877 pKa = 3.37NTFNLSNVLPASVGGKK893 pKa = 8.93SVIEE897 pKa = 4.3DD898 pKa = 3.72LLFDD902 pKa = 3.94KK903 pKa = 11.1VVTSGLGTVDD913 pKa = 3.3TDD915 pKa = 3.77YY916 pKa = 11.21KK917 pKa = 11.24ACAEE921 pKa = 4.05KK922 pKa = 9.97MANTIAEE929 pKa = 4.49AGCVQYY935 pKa = 11.5YY936 pKa = 9.78NGIMVLPGVVDD947 pKa = 3.64QSLLGQYY954 pKa = 7.73TAALTGAMVMGGITAAAAIPFSIAVQSRR982 pKa = 11.84LNYY985 pKa = 10.06LALQTDD991 pKa = 4.07VLQRR995 pKa = 11.84NQQILAQSFNSAMGNITNAFEE1016 pKa = 4.4NVGNAIEE1023 pKa = 4.12QTSQGLQTVAQALDD1037 pKa = 3.47KK1038 pKa = 11.0VQNAVNEE1045 pKa = 4.12QGNALSHH1052 pKa = 5.84LTKK1055 pKa = 10.55QLASNFQAISASIEE1069 pKa = 3.88DD1070 pKa = 4.01LYY1072 pKa = 11.8VRR1074 pKa = 11.84LGQVEE1079 pKa = 4.59ADD1081 pKa = 3.54QQVDD1085 pKa = 3.49RR1086 pKa = 11.84LITGRR1091 pKa = 11.84LAALNAFVSQQLTKK1105 pKa = 9.57YY1106 pKa = 8.06TDD1108 pKa = 3.16VRR1110 pKa = 11.84ASRR1113 pKa = 11.84QLAQEE1118 pKa = 4.75KK1119 pKa = 10.45INEE1122 pKa = 4.44CVKK1125 pKa = 10.63SQSFRR1130 pKa = 11.84YY1131 pKa = 8.66GFCGNGTHH1139 pKa = 5.87VFSVANAAPDD1149 pKa = 4.35GIMFFHH1155 pKa = 7.06SVLLPTAYY1163 pKa = 10.48AEE1165 pKa = 4.25VNAYY1169 pKa = 10.26AGLCVDD1175 pKa = 3.32YY1176 pKa = 10.6KK1177 pKa = 11.25GYY1179 pKa = 10.42VLRR1182 pKa = 11.84DD1183 pKa = 3.44PGNVLFQKK1191 pKa = 10.27PGNEE1195 pKa = 4.26TYY1197 pKa = 10.87LITPRR1202 pKa = 11.84RR1203 pKa = 11.84MFEE1206 pKa = 3.66PRR1208 pKa = 11.84VPQMSDD1214 pKa = 2.99FVQISGCNVSYY1225 pKa = 11.53VNVSSEE1231 pKa = 4.04QLPDD1235 pKa = 3.41IVPDD1239 pKa = 4.35FMDD1242 pKa = 3.64VNATIQDD1249 pKa = 3.82ILDD1252 pKa = 3.66KK1253 pKa = 10.5LQNYY1257 pKa = 7.61SKK1259 pKa = 10.75PDD1261 pKa = 3.56FGLDD1265 pKa = 3.12IFNATYY1271 pKa = 11.05LNLSAEE1277 pKa = 4.48INDD1280 pKa = 3.59LTNRR1284 pKa = 11.84SQQLQAITDD1293 pKa = 3.75EE1294 pKa = 4.24LRR1296 pKa = 11.84ATIANINGTLVDD1308 pKa = 4.67LEE1310 pKa = 4.09WLNRR1314 pKa = 11.84VEE1316 pKa = 5.97TYY1318 pKa = 10.4IKK1320 pKa = 9.3WPWYY1324 pKa = 7.96VWLAIAVALIVLIGLMLWCCLSTGCCGCCSCMASTLDD1361 pKa = 3.63FRR1363 pKa = 11.84GSRR1366 pKa = 11.84LQQYY1370 pKa = 7.6EE1371 pKa = 4.15VEE1373 pKa = 4.46KK1374 pKa = 11.13VHH1376 pKa = 6.56IQQ1378 pKa = 3.09
Molecular weight: 150.49 kDa
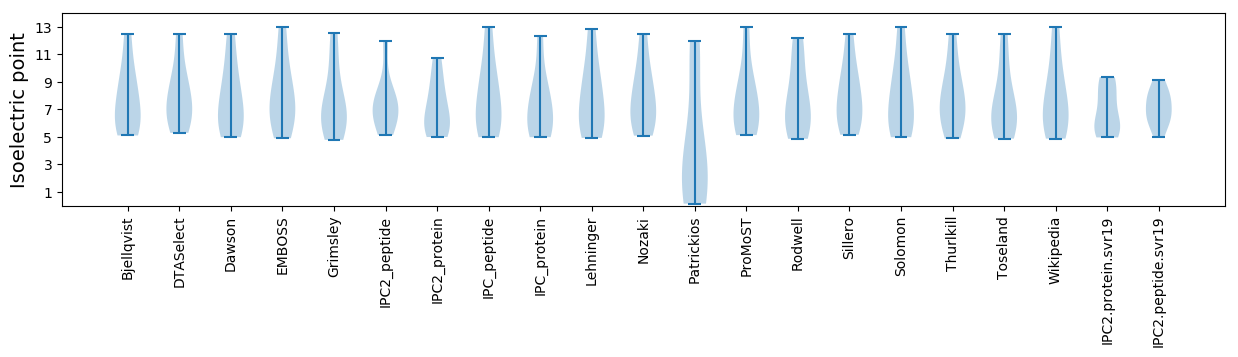
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U1UZ20|A0A0U1UZ20_9ALPC 3C-like proteinase OS=BtMf-AlphaCoV/GD2012 OX=1503280 PE=3 SV=1
MM1 pKa = 7.65FLRR4 pKa = 11.84KK5 pKa = 7.87TRR7 pKa = 11.84TSKK10 pKa = 10.73SSSNRR15 pKa = 11.84LMLIWTLLVRR25 pKa = 11.84TNQRR29 pKa = 11.84SSANLRR35 pKa = 11.84KK36 pKa = 9.72RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 7.23TPLLNSILQHH50 pKa = 6.2LCSHH54 pKa = 7.03HH55 pKa = 6.31LL56 pKa = 3.51
MM1 pKa = 7.65FLRR4 pKa = 11.84KK5 pKa = 7.87TRR7 pKa = 11.84TSKK10 pKa = 10.73SSSNRR15 pKa = 11.84LMLIWTLLVRR25 pKa = 11.84TNQRR29 pKa = 11.84SSANLRR35 pKa = 11.84KK36 pKa = 9.72RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 7.23TPLLNSILQHH50 pKa = 6.2LCSHH54 pKa = 7.03HH55 pKa = 6.31LL56 pKa = 3.51
Molecular weight: 6.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9464 |
56 |
6896 |
1183.0 |
130.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.333 ± 0.228 | 3.318 ± 0.681 |
5.378 ± 0.612 | 3.91 ± 0.372 |
5.811 ± 0.285 | 6.826 ± 0.359 |
1.828 ± 0.347 | 4.522 ± 0.482 |
5.178 ± 0.987 | 8.432 ± 0.919 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.976 ± 0.102 | 5.547 ± 0.737 |
4.068 ± 0.609 | 2.98 ± 0.901 |
3.73 ± 0.656 | 7.069 ± 0.731 |
6.097 ± 0.233 | 10.366 ± 0.741 |
1.205 ± 0.219 | 4.427 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
