
Carsonella ruddii (strain PV)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Zymobacter group; Candidatus Carsonella; Candidatus Carsonella ruddii
Average proteome isoelectric point is 8.91
Get precalculated fractions of proteins
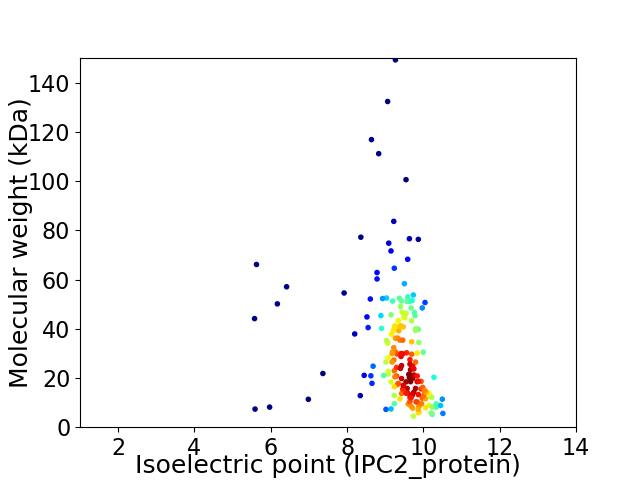
Virtual 2D-PAGE plot for 182 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q05FS9|Q05FS9_CARRP Chaperone protein GrpE OS=Carsonella ruddii (strain PV) OX=387662 GN=CRP_061 PE=3 SV=1
MM1 pKa = 7.4SKK3 pKa = 10.24IIGIDD8 pKa = 3.1LGTTNSCIAVLSNGKK23 pKa = 8.53PQVIEE28 pKa = 3.79NSEE31 pKa = 4.16GGRR34 pKa = 11.84TTPSVVGYY42 pKa = 8.89TEE44 pKa = 4.46DD45 pKa = 3.12NRR47 pKa = 11.84IIVGLPAKK55 pKa = 9.93RR56 pKa = 11.84QAITNPKK63 pKa = 7.39NTLYY67 pKa = 10.75AIKK70 pKa = 10.29RR71 pKa = 11.84LIGRR75 pKa = 11.84KK76 pKa = 9.15FKK78 pKa = 11.12DD79 pKa = 4.33DD80 pKa = 3.5IVQKK84 pKa = 9.98DD85 pKa = 3.58IKK87 pKa = 9.43MVPYY91 pKa = 10.35KK92 pKa = 10.44IISSEE97 pKa = 4.08NGDD100 pKa = 2.84AWVEE104 pKa = 4.19VKK106 pKa = 10.36DD107 pKa = 4.43KK108 pKa = 11.09KK109 pKa = 10.54LAPPQISAEE118 pKa = 4.04ILKK121 pKa = 10.5KK122 pKa = 10.33MKK124 pKa = 8.91ITAEE128 pKa = 3.74NFLNEE133 pKa = 4.34KK134 pKa = 7.18VTKK137 pKa = 10.52AVITVPAYY145 pKa = 10.64FNDD148 pKa = 3.76SQRR151 pKa = 11.84QATKK155 pKa = 10.65DD156 pKa = 3.08AGKK159 pKa = 10.12IAGLEE164 pKa = 3.77VLRR167 pKa = 11.84IINEE171 pKa = 3.8PTAAALAYY179 pKa = 10.48GLDD182 pKa = 3.31KK183 pKa = 10.93KK184 pKa = 11.32KK185 pKa = 10.42NDD187 pKa = 4.34RR188 pKa = 11.84IIAVYY193 pKa = 10.39DD194 pKa = 3.67LGGGTFDD201 pKa = 4.02ISIIEE206 pKa = 4.03IANVDD211 pKa = 3.71GEE213 pKa = 4.74TQFEE217 pKa = 4.31VLSTNGDD224 pKa = 3.61TFLGGEE230 pKa = 4.53DD231 pKa = 3.37FDD233 pKa = 5.0IRR235 pKa = 11.84IINNLIYY242 pKa = 10.23EE243 pKa = 4.32FKK245 pKa = 10.47IEE247 pKa = 4.07NGINLSGDD255 pKa = 3.57SLAMQRR261 pKa = 11.84LKK263 pKa = 10.8EE264 pKa = 3.98AAEE267 pKa = 3.95KK268 pKa = 10.88AKK270 pKa = 10.53IEE272 pKa = 4.1LSSVEE277 pKa = 3.85QTDD280 pKa = 2.98INLPYY285 pKa = 9.34ITADD289 pKa = 3.2KK290 pKa = 10.96NGPKK294 pKa = 9.93HH295 pKa = 6.49LNIKK299 pKa = 8.1ITRR302 pKa = 11.84SKK304 pKa = 11.05LEE306 pKa = 3.96SLVEE310 pKa = 3.86DD311 pKa = 5.76LILKK315 pKa = 8.96SLKK318 pKa = 9.89PCEE321 pKa = 3.96IALNDD326 pKa = 4.18AKK328 pKa = 10.6ISKK331 pKa = 10.29NKK333 pKa = 8.79IDD335 pKa = 4.33EE336 pKa = 4.83IILVGGQTRR345 pKa = 11.84MPLVQKK351 pKa = 9.29MVSDD355 pKa = 4.12FFEE358 pKa = 4.45KK359 pKa = 10.62VVKK362 pKa = 10.48KK363 pKa = 10.61DD364 pKa = 3.42INPDD368 pKa = 3.02EE369 pKa = 4.32AVAIGASVQAGVLSGVVKK387 pKa = 10.69DD388 pKa = 4.1VLLLDD393 pKa = 3.83VTPLTLGIEE402 pKa = 4.34TMGGIMTPLIEE413 pKa = 4.93KK414 pKa = 8.43NTTIPTKK421 pKa = 9.18KK422 pKa = 7.9TQVFSTAEE430 pKa = 4.01DD431 pKa = 3.57NQTSVTIHH439 pKa = 5.96TLQGEE444 pKa = 4.46RR445 pKa = 11.84KK446 pKa = 9.26KK447 pKa = 10.88ALQNKK452 pKa = 9.55SLGKK456 pKa = 10.07FDD458 pKa = 5.67LNNISPAPRR467 pKa = 11.84GVPQIEE473 pKa = 4.08VSFDD477 pKa = 3.25LDD479 pKa = 3.65ANGILNVTAKK489 pKa = 10.49DD490 pKa = 3.16KK491 pKa = 10.47KK492 pKa = 9.18TGVEE496 pKa = 3.56QSIVIKK502 pKa = 10.81SSGGLSEE509 pKa = 5.73LEE511 pKa = 3.93IEE513 pKa = 5.0NMIKK517 pKa = 10.47DD518 pKa = 3.68AEE520 pKa = 4.39ANLEE524 pKa = 3.78IDD526 pKa = 3.88KK527 pKa = 11.08KK528 pKa = 11.0FEE530 pKa = 4.0EE531 pKa = 4.44LVKK534 pKa = 10.7CRR536 pKa = 11.84NEE538 pKa = 3.82ADD540 pKa = 3.56STISIVKK547 pKa = 10.26KK548 pKa = 10.29KK549 pKa = 10.99LKK551 pKa = 10.59DD552 pKa = 3.63EE553 pKa = 4.05NLKK556 pKa = 10.42ILDD559 pKa = 3.9EE560 pKa = 4.24EE561 pKa = 4.26RR562 pKa = 11.84VSIEE566 pKa = 3.66KK567 pKa = 10.58SISNLEE573 pKa = 3.99LLIKK577 pKa = 10.64GDD579 pKa = 5.22DD580 pKa = 3.24IDD582 pKa = 4.89SIKK585 pKa = 10.81KK586 pKa = 10.04EE587 pKa = 3.96NEE589 pKa = 3.76EE590 pKa = 4.27LLKK593 pKa = 11.25LSDD596 pKa = 4.31NIIKK600 pKa = 10.48KK601 pKa = 10.01KK602 pKa = 10.47
MM1 pKa = 7.4SKK3 pKa = 10.24IIGIDD8 pKa = 3.1LGTTNSCIAVLSNGKK23 pKa = 8.53PQVIEE28 pKa = 3.79NSEE31 pKa = 4.16GGRR34 pKa = 11.84TTPSVVGYY42 pKa = 8.89TEE44 pKa = 4.46DD45 pKa = 3.12NRR47 pKa = 11.84IIVGLPAKK55 pKa = 9.93RR56 pKa = 11.84QAITNPKK63 pKa = 7.39NTLYY67 pKa = 10.75AIKK70 pKa = 10.29RR71 pKa = 11.84LIGRR75 pKa = 11.84KK76 pKa = 9.15FKK78 pKa = 11.12DD79 pKa = 4.33DD80 pKa = 3.5IVQKK84 pKa = 9.98DD85 pKa = 3.58IKK87 pKa = 9.43MVPYY91 pKa = 10.35KK92 pKa = 10.44IISSEE97 pKa = 4.08NGDD100 pKa = 2.84AWVEE104 pKa = 4.19VKK106 pKa = 10.36DD107 pKa = 4.43KK108 pKa = 11.09KK109 pKa = 10.54LAPPQISAEE118 pKa = 4.04ILKK121 pKa = 10.5KK122 pKa = 10.33MKK124 pKa = 8.91ITAEE128 pKa = 3.74NFLNEE133 pKa = 4.34KK134 pKa = 7.18VTKK137 pKa = 10.52AVITVPAYY145 pKa = 10.64FNDD148 pKa = 3.76SQRR151 pKa = 11.84QATKK155 pKa = 10.65DD156 pKa = 3.08AGKK159 pKa = 10.12IAGLEE164 pKa = 3.77VLRR167 pKa = 11.84IINEE171 pKa = 3.8PTAAALAYY179 pKa = 10.48GLDD182 pKa = 3.31KK183 pKa = 10.93KK184 pKa = 11.32KK185 pKa = 10.42NDD187 pKa = 4.34RR188 pKa = 11.84IIAVYY193 pKa = 10.39DD194 pKa = 3.67LGGGTFDD201 pKa = 4.02ISIIEE206 pKa = 4.03IANVDD211 pKa = 3.71GEE213 pKa = 4.74TQFEE217 pKa = 4.31VLSTNGDD224 pKa = 3.61TFLGGEE230 pKa = 4.53DD231 pKa = 3.37FDD233 pKa = 5.0IRR235 pKa = 11.84IINNLIYY242 pKa = 10.23EE243 pKa = 4.32FKK245 pKa = 10.47IEE247 pKa = 4.07NGINLSGDD255 pKa = 3.57SLAMQRR261 pKa = 11.84LKK263 pKa = 10.8EE264 pKa = 3.98AAEE267 pKa = 3.95KK268 pKa = 10.88AKK270 pKa = 10.53IEE272 pKa = 4.1LSSVEE277 pKa = 3.85QTDD280 pKa = 2.98INLPYY285 pKa = 9.34ITADD289 pKa = 3.2KK290 pKa = 10.96NGPKK294 pKa = 9.93HH295 pKa = 6.49LNIKK299 pKa = 8.1ITRR302 pKa = 11.84SKK304 pKa = 11.05LEE306 pKa = 3.96SLVEE310 pKa = 3.86DD311 pKa = 5.76LILKK315 pKa = 8.96SLKK318 pKa = 9.89PCEE321 pKa = 3.96IALNDD326 pKa = 4.18AKK328 pKa = 10.6ISKK331 pKa = 10.29NKK333 pKa = 8.79IDD335 pKa = 4.33EE336 pKa = 4.83IILVGGQTRR345 pKa = 11.84MPLVQKK351 pKa = 9.29MVSDD355 pKa = 4.12FFEE358 pKa = 4.45KK359 pKa = 10.62VVKK362 pKa = 10.48KK363 pKa = 10.61DD364 pKa = 3.42INPDD368 pKa = 3.02EE369 pKa = 4.32AVAIGASVQAGVLSGVVKK387 pKa = 10.69DD388 pKa = 4.1VLLLDD393 pKa = 3.83VTPLTLGIEE402 pKa = 4.34TMGGIMTPLIEE413 pKa = 4.93KK414 pKa = 8.43NTTIPTKK421 pKa = 9.18KK422 pKa = 7.9TQVFSTAEE430 pKa = 4.01DD431 pKa = 3.57NQTSVTIHH439 pKa = 5.96TLQGEE444 pKa = 4.46RR445 pKa = 11.84KK446 pKa = 9.26KK447 pKa = 10.88ALQNKK452 pKa = 9.55SLGKK456 pKa = 10.07FDD458 pKa = 5.67LNNISPAPRR467 pKa = 11.84GVPQIEE473 pKa = 4.08VSFDD477 pKa = 3.25LDD479 pKa = 3.65ANGILNVTAKK489 pKa = 10.49DD490 pKa = 3.16KK491 pKa = 10.47KK492 pKa = 9.18TGVEE496 pKa = 3.56QSIVIKK502 pKa = 10.81SSGGLSEE509 pKa = 5.73LEE511 pKa = 3.93IEE513 pKa = 5.0NMIKK517 pKa = 10.47DD518 pKa = 3.68AEE520 pKa = 4.39ANLEE524 pKa = 3.78IDD526 pKa = 3.88KK527 pKa = 11.08KK528 pKa = 11.0FEE530 pKa = 4.0EE531 pKa = 4.44LVKK534 pKa = 10.7CRR536 pKa = 11.84NEE538 pKa = 3.82ADD540 pKa = 3.56STISIVKK547 pKa = 10.26KK548 pKa = 10.29KK549 pKa = 10.99LKK551 pKa = 10.59DD552 pKa = 3.63EE553 pKa = 4.05NLKK556 pKa = 10.42ILDD559 pKa = 3.9EE560 pKa = 4.24EE561 pKa = 4.26RR562 pKa = 11.84VSIEE566 pKa = 3.66KK567 pKa = 10.58SISNLEE573 pKa = 3.99LLIKK577 pKa = 10.64GDD579 pKa = 5.22DD580 pKa = 3.24IDD582 pKa = 4.89SIKK585 pKa = 10.81KK586 pKa = 10.04EE587 pKa = 3.96NEE589 pKa = 3.76EE590 pKa = 4.27LLKK593 pKa = 11.25LSDD596 pKa = 4.31NIIKK600 pKa = 10.48KK601 pKa = 10.01KK602 pKa = 10.47
Molecular weight: 66.12 kDa
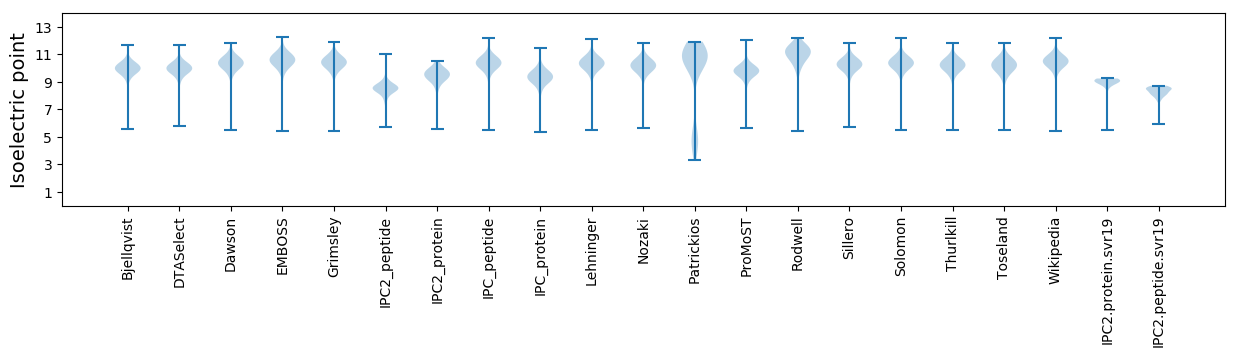
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q05FI1|RS7_CARRP 30S ribosomal protein S7 OS=Carsonella ruddii (strain PV) OX=387662 GN=rpsG PE=3 SV=1
MM1 pKa = 7.58TLNQILKK8 pKa = 9.49FKK10 pKa = 10.34RR11 pKa = 11.84KK12 pKa = 9.85KK13 pKa = 9.94SVKK16 pKa = 9.63KK17 pKa = 10.43KK18 pKa = 8.99KK19 pKa = 8.94TPALLSSPQKK29 pKa = 10.51KK30 pKa = 10.01GICIKK35 pKa = 10.84VYY37 pKa = 7.54TTTPKK42 pKa = 10.53KK43 pKa = 10.26PNSALRR49 pKa = 11.84KK50 pKa = 7.35VCRR53 pKa = 11.84VKK55 pKa = 10.92LSNKK59 pKa = 9.92NEE61 pKa = 3.43ITAYY65 pKa = 9.83IPGEE69 pKa = 3.86GHH71 pKa = 6.66NLQEE75 pKa = 4.3HH76 pKa = 6.37SNVLVRR82 pKa = 11.84GGRR85 pKa = 11.84VKK87 pKa = 10.82DD88 pKa = 3.7LPGVKK93 pKa = 9.6YY94 pKa = 10.19HH95 pKa = 7.61IIRR98 pKa = 11.84NVYY101 pKa = 9.84DD102 pKa = 3.63LSGVINRR109 pKa = 11.84KK110 pKa = 7.0TSRR113 pKa = 11.84SKK115 pKa = 10.87YY116 pKa = 9.16GKK118 pKa = 10.17KK119 pKa = 9.9KK120 pKa = 10.55
MM1 pKa = 7.58TLNQILKK8 pKa = 9.49FKK10 pKa = 10.34RR11 pKa = 11.84KK12 pKa = 9.85KK13 pKa = 9.94SVKK16 pKa = 9.63KK17 pKa = 10.43KK18 pKa = 8.99KK19 pKa = 8.94TPALLSSPQKK29 pKa = 10.51KK30 pKa = 10.01GICIKK35 pKa = 10.84VYY37 pKa = 7.54TTTPKK42 pKa = 10.53KK43 pKa = 10.26PNSALRR49 pKa = 11.84KK50 pKa = 7.35VCRR53 pKa = 11.84VKK55 pKa = 10.92LSNKK59 pKa = 9.92NEE61 pKa = 3.43ITAYY65 pKa = 9.83IPGEE69 pKa = 3.86GHH71 pKa = 6.66NLQEE75 pKa = 4.3HH76 pKa = 6.37SNVLVRR82 pKa = 11.84GGRR85 pKa = 11.84VKK87 pKa = 10.82DD88 pKa = 3.7LPGVKK93 pKa = 9.6YY94 pKa = 10.19HH95 pKa = 7.61IIRR98 pKa = 11.84NVYY101 pKa = 9.84DD102 pKa = 3.63LSGVINRR109 pKa = 11.84KK110 pKa = 7.0TSRR113 pKa = 11.84SKK115 pKa = 10.87YY116 pKa = 9.16GKK118 pKa = 10.17KK119 pKa = 9.9KK120 pKa = 10.55
Molecular weight: 13.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
49923 |
37 |
1292 |
274.3 |
32.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.478 ± 0.135 | 1.633 ± 0.063 |
2.62 ± 0.115 | 3.327 ± 0.157 |
8.994 ± 0.251 | 3.698 ± 0.179 |
1.054 ± 0.049 | 15.522 ± 0.198 |
14.799 ± 0.35 | 10.134 ± 0.151 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.238 ± 0.057 | 11.776 ± 0.233 |
1.62 ± 0.08 | 1.476 ± 0.066 |
1.857 ± 0.09 | 6.344 ± 0.156 |
3.177 ± 0.102 | 3.219 ± 0.123 |
0.463 ± 0.049 | 5.573 ± 0.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
