
Abalone herpesvirus (isolate Abalone/Australia/Victoria/2009) (AbHV)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Malacoherpesviridae; Aurivirus; Haliotid herpesvirus 1
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
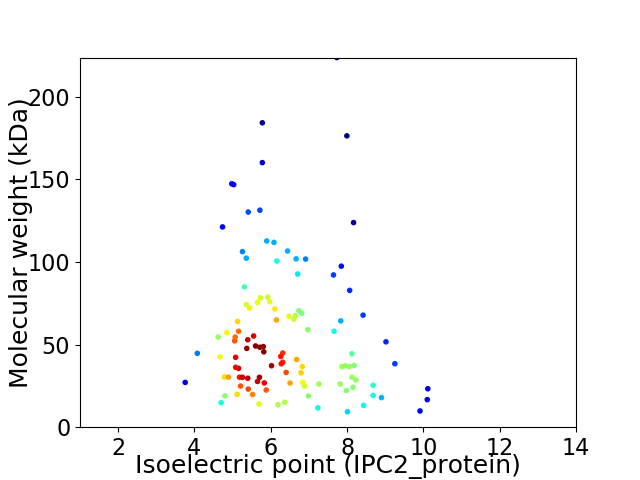
Virtual 2D-PAGE plot for 112 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4JUE5|K4JUE5_ABHV Uncharacterized protein OS=Abalone herpesvirus (isolate Abalone/Australia/Victoria/2009) OX=1241371 GN=AbHV_ORF19 PE=4 SV=1
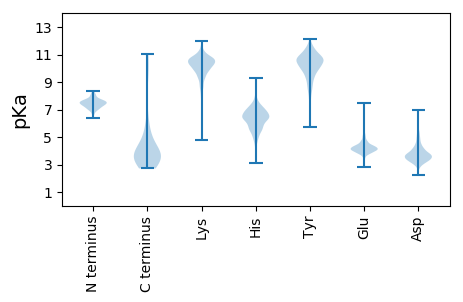
MM1 pKa = 8.12DD2 pKa = 5.86DD3 pKa = 4.46KK4 pKa = 11.63SSGMQAAALATEE16 pKa = 4.94DD17 pKa = 5.05DD18 pKa = 4.21DD19 pKa = 6.99DD20 pKa = 4.74GVSSLVDD27 pKa = 3.94MINSGALSLEE37 pKa = 4.52VEE39 pKa = 4.34RR40 pKa = 11.84QMKK43 pKa = 10.21RR44 pKa = 11.84DD45 pKa = 3.42GEE47 pKa = 4.38RR48 pKa = 11.84NLLFYY53 pKa = 10.89QEE55 pKa = 4.14VMSATFDD62 pKa = 3.83RR63 pKa = 11.84DD64 pKa = 3.32LLRR67 pKa = 11.84SKK69 pKa = 10.41IFPEE73 pKa = 4.49DD74 pKa = 3.58ASKK77 pKa = 10.65GPCMRR82 pKa = 11.84MKK84 pKa = 10.47QSLYY88 pKa = 10.81KK89 pKa = 9.99EE90 pKa = 3.82ASKK93 pKa = 10.64PIVGEE98 pKa = 4.0YY99 pKa = 10.57AFEE102 pKa = 4.47SDD104 pKa = 4.25PVFSSASTAHH114 pKa = 6.12LRR116 pKa = 11.84LVEE119 pKa = 3.96KK120 pKa = 10.85SKK122 pKa = 10.73ILSKK126 pKa = 10.54DD127 pKa = 2.77IPEE130 pKa = 4.22IVLDD134 pKa = 4.04YY135 pKa = 11.11NQDD138 pKa = 3.35PSVISVDD145 pKa = 3.59EE146 pKa = 4.44SNMIVTDD153 pKa = 3.16VHH155 pKa = 6.87HH156 pKa = 6.42PQEE159 pKa = 4.11NQTRR163 pKa = 11.84IFFQLDD169 pKa = 2.9NGYY172 pKa = 9.6VLSIPTVNPWSEE184 pKa = 3.69RR185 pKa = 11.84IYY187 pKa = 10.32RR188 pKa = 11.84YY189 pKa = 8.64TITSVPQDD197 pKa = 3.66KK198 pKa = 10.65EE199 pKa = 3.93LLVYY203 pKa = 10.92DD204 pKa = 3.9NLEE207 pKa = 4.24DD208 pKa = 4.06VFKK211 pKa = 10.96NYY213 pKa = 10.06PNYY216 pKa = 10.56AEE218 pKa = 4.95KK219 pKa = 10.96LFTNAPDD226 pKa = 5.4DD227 pKa = 5.31DD228 pKa = 6.56DD229 pKa = 6.93DD230 pKa = 6.33DD231 pKa = 5.21EE232 pKa = 5.37EE233 pKa = 5.64GGPEE237 pKa = 4.1QYY239 pKa = 11.09LKK241 pKa = 10.85LDD243 pKa = 3.5RR244 pKa = 11.84SFLGVSGVSDD254 pKa = 3.89GSDD257 pKa = 3.36LEE259 pKa = 4.68VVARR263 pKa = 11.84YY264 pKa = 8.45HH265 pKa = 7.5IPMEE269 pKa = 4.61LNPLQYY275 pKa = 11.29VMHH278 pKa = 6.97KK279 pKa = 10.01PNLYY283 pKa = 10.53LFTGKK288 pKa = 9.97DD289 pKa = 3.22QEE291 pKa = 4.6GNNNCLYY298 pKa = 11.01ALNTEE303 pKa = 4.24LVEE306 pKa = 4.73GEE308 pKa = 4.38GKK310 pKa = 9.59PWIAPWTKK318 pKa = 10.14VKK320 pKa = 10.32IYY322 pKa = 10.65DD323 pKa = 3.47SEE325 pKa = 4.95TYY327 pKa = 10.94VDD329 pKa = 4.01LTDD332 pKa = 4.72PNTLTVFEE340 pKa = 5.44DD341 pKa = 3.93EE342 pKa = 4.55VPSYY346 pKa = 11.06RR347 pKa = 11.84LTIEE351 pKa = 3.76VSEE354 pKa = 4.29GSNIGVLSVPDD365 pKa = 3.91YY366 pKa = 7.96PTYY369 pKa = 10.46DD370 pKa = 3.41IPSLTLIEE378 pKa = 4.23EE379 pKa = 4.29VNDD382 pKa = 3.5IVLEE386 pKa = 4.16AAEE389 pKa = 4.64EE390 pKa = 4.07EE391 pKa = 4.35DD392 pKa = 6.18DD393 pKa = 4.39EE394 pKa = 5.26DD395 pKa = 4.15
MM1 pKa = 8.12DD2 pKa = 5.86DD3 pKa = 4.46KK4 pKa = 11.63SSGMQAAALATEE16 pKa = 4.94DD17 pKa = 5.05DD18 pKa = 4.21DD19 pKa = 6.99DD20 pKa = 4.74GVSSLVDD27 pKa = 3.94MINSGALSLEE37 pKa = 4.52VEE39 pKa = 4.34RR40 pKa = 11.84QMKK43 pKa = 10.21RR44 pKa = 11.84DD45 pKa = 3.42GEE47 pKa = 4.38RR48 pKa = 11.84NLLFYY53 pKa = 10.89QEE55 pKa = 4.14VMSATFDD62 pKa = 3.83RR63 pKa = 11.84DD64 pKa = 3.32LLRR67 pKa = 11.84SKK69 pKa = 10.41IFPEE73 pKa = 4.49DD74 pKa = 3.58ASKK77 pKa = 10.65GPCMRR82 pKa = 11.84MKK84 pKa = 10.47QSLYY88 pKa = 10.81KK89 pKa = 9.99EE90 pKa = 3.82ASKK93 pKa = 10.64PIVGEE98 pKa = 4.0YY99 pKa = 10.57AFEE102 pKa = 4.47SDD104 pKa = 4.25PVFSSASTAHH114 pKa = 6.12LRR116 pKa = 11.84LVEE119 pKa = 3.96KK120 pKa = 10.85SKK122 pKa = 10.73ILSKK126 pKa = 10.54DD127 pKa = 2.77IPEE130 pKa = 4.22IVLDD134 pKa = 4.04YY135 pKa = 11.11NQDD138 pKa = 3.35PSVISVDD145 pKa = 3.59EE146 pKa = 4.44SNMIVTDD153 pKa = 3.16VHH155 pKa = 6.87HH156 pKa = 6.42PQEE159 pKa = 4.11NQTRR163 pKa = 11.84IFFQLDD169 pKa = 2.9NGYY172 pKa = 9.6VLSIPTVNPWSEE184 pKa = 3.69RR185 pKa = 11.84IYY187 pKa = 10.32RR188 pKa = 11.84YY189 pKa = 8.64TITSVPQDD197 pKa = 3.66KK198 pKa = 10.65EE199 pKa = 3.93LLVYY203 pKa = 10.92DD204 pKa = 3.9NLEE207 pKa = 4.24DD208 pKa = 4.06VFKK211 pKa = 10.96NYY213 pKa = 10.06PNYY216 pKa = 10.56AEE218 pKa = 4.95KK219 pKa = 10.96LFTNAPDD226 pKa = 5.4DD227 pKa = 5.31DD228 pKa = 6.56DD229 pKa = 6.93DD230 pKa = 6.33DD231 pKa = 5.21EE232 pKa = 5.37EE233 pKa = 5.64GGPEE237 pKa = 4.1QYY239 pKa = 11.09LKK241 pKa = 10.85LDD243 pKa = 3.5RR244 pKa = 11.84SFLGVSGVSDD254 pKa = 3.89GSDD257 pKa = 3.36LEE259 pKa = 4.68VVARR263 pKa = 11.84YY264 pKa = 8.45HH265 pKa = 7.5IPMEE269 pKa = 4.61LNPLQYY275 pKa = 11.29VMHH278 pKa = 6.97KK279 pKa = 10.01PNLYY283 pKa = 10.53LFTGKK288 pKa = 9.97DD289 pKa = 3.22QEE291 pKa = 4.6GNNNCLYY298 pKa = 11.01ALNTEE303 pKa = 4.24LVEE306 pKa = 4.73GEE308 pKa = 4.38GKK310 pKa = 9.59PWIAPWTKK318 pKa = 10.14VKK320 pKa = 10.32IYY322 pKa = 10.65DD323 pKa = 3.47SEE325 pKa = 4.95TYY327 pKa = 10.94VDD329 pKa = 4.01LTDD332 pKa = 4.72PNTLTVFEE340 pKa = 5.44DD341 pKa = 3.93EE342 pKa = 4.55VPSYY346 pKa = 11.06RR347 pKa = 11.84LTIEE351 pKa = 3.76VSEE354 pKa = 4.29GSNIGVLSVPDD365 pKa = 3.91YY366 pKa = 7.96PTYY369 pKa = 10.46DD370 pKa = 3.41IPSLTLIEE378 pKa = 4.23EE379 pKa = 4.29VNDD382 pKa = 3.5IVLEE386 pKa = 4.16AAEE389 pKa = 4.64EE390 pKa = 4.07EE391 pKa = 4.35DD392 pKa = 6.18DD393 pKa = 4.39EE394 pKa = 5.26DD395 pKa = 4.15
Molecular weight: 44.68 kDa
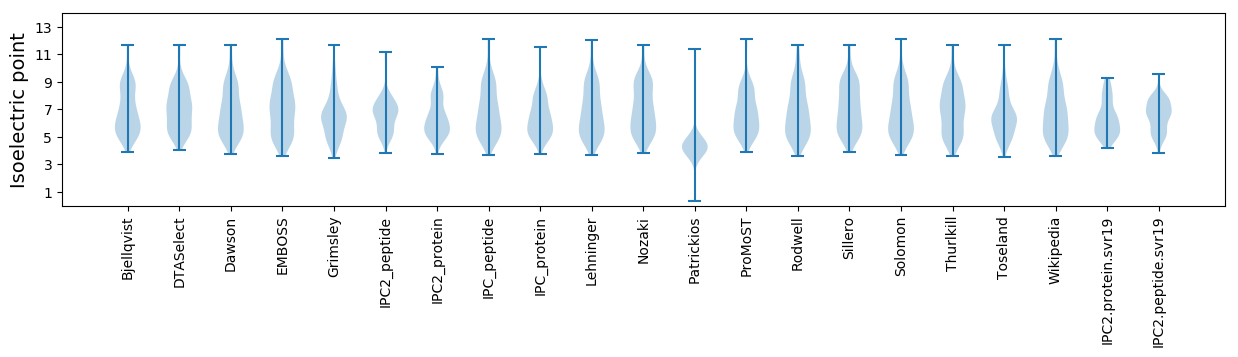
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4JYI3|K4JYI3_ABHV Uncharacterized protein OS=Abalone herpesvirus (isolate Abalone/Australia/Victoria/2009) OX=1241371 GN=AbHV_ORF66 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.29FVTRR6 pKa = 11.84KK7 pKa = 10.0RR8 pKa = 11.84LVTKK12 pKa = 10.14MLLTHH17 pKa = 6.76KK18 pKa = 10.07FKK20 pKa = 11.12LVTRR24 pKa = 11.84TGLATKK30 pKa = 10.11KK31 pKa = 10.61YY32 pKa = 10.1LALKK36 pKa = 10.12MNDD39 pKa = 2.99TWGKK43 pKa = 10.09DD44 pKa = 3.15ALSRR48 pKa = 11.84AEE50 pKa = 3.61INYY53 pKa = 9.61VNEE56 pKa = 3.57NDD58 pKa = 3.5TWGKK62 pKa = 9.24NAPTLQKK69 pKa = 10.41NLRR72 pKa = 11.84SAVFLVPTSIVHH84 pKa = 5.75AQHH87 pKa = 6.46PQRR90 pKa = 11.84KK91 pKa = 7.15RR92 pKa = 11.84TEE94 pKa = 3.71ARR96 pKa = 11.84LPPSLNLQRR105 pKa = 11.84TLTDD109 pKa = 2.82SKK111 pKa = 10.3MYY113 pKa = 9.94PSLKK117 pKa = 9.78IILRR121 pKa = 11.84RR122 pKa = 11.84KK123 pKa = 8.31RR124 pKa = 11.84TVATLLPSLKK134 pKa = 10.12KK135 pKa = 10.51KK136 pKa = 10.21LWSGLCWTT144 pKa = 5.04
MM1 pKa = 7.47KK2 pKa = 10.29FVTRR6 pKa = 11.84KK7 pKa = 10.0RR8 pKa = 11.84LVTKK12 pKa = 10.14MLLTHH17 pKa = 6.76KK18 pKa = 10.07FKK20 pKa = 11.12LVTRR24 pKa = 11.84TGLATKK30 pKa = 10.11KK31 pKa = 10.61YY32 pKa = 10.1LALKK36 pKa = 10.12MNDD39 pKa = 2.99TWGKK43 pKa = 10.09DD44 pKa = 3.15ALSRR48 pKa = 11.84AEE50 pKa = 3.61INYY53 pKa = 9.61VNEE56 pKa = 3.57NDD58 pKa = 3.5TWGKK62 pKa = 9.24NAPTLQKK69 pKa = 10.41NLRR72 pKa = 11.84SAVFLVPTSIVHH84 pKa = 5.75AQHH87 pKa = 6.46PQRR90 pKa = 11.84KK91 pKa = 7.15RR92 pKa = 11.84TEE94 pKa = 3.71ARR96 pKa = 11.84LPPSLNLQRR105 pKa = 11.84TLTDD109 pKa = 2.82SKK111 pKa = 10.3MYY113 pKa = 9.94PSLKK117 pKa = 9.78IILRR121 pKa = 11.84RR122 pKa = 11.84KK123 pKa = 8.31RR124 pKa = 11.84TVATLLPSLKK134 pKa = 10.12KK135 pKa = 10.51KK136 pKa = 10.21LWSGLCWTT144 pKa = 5.04
Molecular weight: 16.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
56122 |
79 |
1926 |
501.1 |
57.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.7 ± 0.13 | 1.937 ± 0.118 |
5.928 ± 0.13 | 8.025 ± 0.247 |
4.321 ± 0.095 | 4.994 ± 0.12 |
1.994 ± 0.101 | 5.675 ± 0.126 |
7.658 ± 0.196 | 8.269 ± 0.195 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.067 ± 0.096 | 5.023 ± 0.119 |
4.572 ± 0.18 | 3.304 ± 0.09 |
5.399 ± 0.168 | 6.976 ± 0.15 |
5.958 ± 0.144 | 6.971 ± 0.1 |
0.798 ± 0.043 | 3.43 ± 0.146 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
