
Faeces associated gemycircularvirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus geras3; Gerygone associated gemycircularvirus 3
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
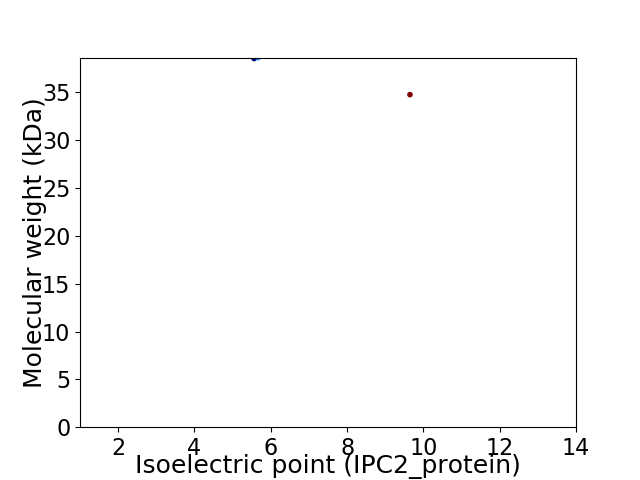
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YTC0|T1YTC0_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 3 OX=1391032 PE=4 SV=1
MM1 pKa = 7.55PPKK4 pKa = 10.38AKK6 pKa = 9.95PNPYY10 pKa = 9.72FADD13 pKa = 3.11GRR15 pKa = 11.84YY16 pKa = 9.19FLLTYY21 pKa = 8.77AQCGTLDD28 pKa = 2.95AWTVNDD34 pKa = 3.97HH35 pKa = 6.64LAFLGAEE42 pKa = 4.25CIIGRR47 pKa = 11.84EE48 pKa = 4.02LHH50 pKa = 6.83ADD52 pKa = 3.72GGTHH56 pKa = 6.78LHH58 pKa = 6.66AFCDD62 pKa = 5.08FSRR65 pKa = 11.84RR66 pKa = 11.84FRR68 pKa = 11.84SRR70 pKa = 11.84RR71 pKa = 11.84SDD73 pKa = 3.3VFDD76 pKa = 3.2VGGRR80 pKa = 11.84HH81 pKa = 6.27PNLVPSYY88 pKa = 8.82GKK90 pKa = 9.76PEE92 pKa = 3.92GGYY95 pKa = 10.65DD96 pKa = 3.55YY97 pKa = 10.74AIKK100 pKa = 10.94DD101 pKa = 3.59GDD103 pKa = 4.03VVAGGLSRR111 pKa = 11.84PSGRR115 pKa = 11.84GVYY118 pKa = 10.06EE119 pKa = 4.89DD120 pKa = 3.54VSSWSIIVGQEE131 pKa = 3.87SEE133 pKa = 4.58GEE135 pKa = 3.92FWKK138 pKa = 10.87CVARR142 pKa = 11.84LDD144 pKa = 4.55PRR146 pKa = 11.84ALCTNYY152 pKa = 10.56NSLRR156 pKa = 11.84AYY158 pKa = 10.43ANWRR162 pKa = 11.84YY163 pKa = 9.76RR164 pKa = 11.84PAPVPYY170 pKa = 8.89EE171 pKa = 4.04HH172 pKa = 7.45PAGIEE177 pKa = 4.08FEE179 pKa = 4.46LGMVPEE185 pKa = 4.27LAVWRR190 pKa = 11.84EE191 pKa = 3.56IALGADD197 pKa = 3.77RR198 pKa = 11.84EE199 pKa = 4.16RR200 pKa = 11.84GILVIFGDD208 pKa = 3.67TRR210 pKa = 11.84LGKK213 pKa = 7.65TLWARR218 pKa = 11.84SIGPHH223 pKa = 6.89IYY225 pKa = 10.2TIGQMSGEE233 pKa = 4.38VILRR237 pKa = 11.84DD238 pKa = 4.51GPDD241 pKa = 2.84ADD243 pKa = 3.79YY244 pKa = 11.53AVFDD248 pKa = 4.39DD249 pKa = 4.11MRR251 pKa = 11.84GGLEE255 pKa = 4.55FFHH258 pKa = 6.92GWKK261 pKa = 9.71EE262 pKa = 3.86WFGCQSVVTVKK273 pKa = 10.58KK274 pKa = 10.58LYY276 pKa = 9.92RR277 pKa = 11.84DD278 pKa = 3.77PVQMPWGKK286 pKa = 9.33PVIWLSNRR294 pKa = 11.84DD295 pKa = 3.57PRR297 pKa = 11.84DD298 pKa = 3.33EE299 pKa = 4.41LRR301 pKa = 11.84DD302 pKa = 3.78SITNHH307 pKa = 4.96TSMGKK312 pKa = 8.57QASIEE317 pKa = 4.03GDD319 pKa = 3.59IKK321 pKa = 10.77WLDD324 pKa = 3.56GNCIFVEE331 pKa = 4.06LDD333 pKa = 3.15HH334 pKa = 7.64AIFRR338 pKa = 11.84ANTEE342 pKa = 3.84
MM1 pKa = 7.55PPKK4 pKa = 10.38AKK6 pKa = 9.95PNPYY10 pKa = 9.72FADD13 pKa = 3.11GRR15 pKa = 11.84YY16 pKa = 9.19FLLTYY21 pKa = 8.77AQCGTLDD28 pKa = 2.95AWTVNDD34 pKa = 3.97HH35 pKa = 6.64LAFLGAEE42 pKa = 4.25CIIGRR47 pKa = 11.84EE48 pKa = 4.02LHH50 pKa = 6.83ADD52 pKa = 3.72GGTHH56 pKa = 6.78LHH58 pKa = 6.66AFCDD62 pKa = 5.08FSRR65 pKa = 11.84RR66 pKa = 11.84FRR68 pKa = 11.84SRR70 pKa = 11.84RR71 pKa = 11.84SDD73 pKa = 3.3VFDD76 pKa = 3.2VGGRR80 pKa = 11.84HH81 pKa = 6.27PNLVPSYY88 pKa = 8.82GKK90 pKa = 9.76PEE92 pKa = 3.92GGYY95 pKa = 10.65DD96 pKa = 3.55YY97 pKa = 10.74AIKK100 pKa = 10.94DD101 pKa = 3.59GDD103 pKa = 4.03VVAGGLSRR111 pKa = 11.84PSGRR115 pKa = 11.84GVYY118 pKa = 10.06EE119 pKa = 4.89DD120 pKa = 3.54VSSWSIIVGQEE131 pKa = 3.87SEE133 pKa = 4.58GEE135 pKa = 3.92FWKK138 pKa = 10.87CVARR142 pKa = 11.84LDD144 pKa = 4.55PRR146 pKa = 11.84ALCTNYY152 pKa = 10.56NSLRR156 pKa = 11.84AYY158 pKa = 10.43ANWRR162 pKa = 11.84YY163 pKa = 9.76RR164 pKa = 11.84PAPVPYY170 pKa = 8.89EE171 pKa = 4.04HH172 pKa = 7.45PAGIEE177 pKa = 4.08FEE179 pKa = 4.46LGMVPEE185 pKa = 4.27LAVWRR190 pKa = 11.84EE191 pKa = 3.56IALGADD197 pKa = 3.77RR198 pKa = 11.84EE199 pKa = 4.16RR200 pKa = 11.84GILVIFGDD208 pKa = 3.67TRR210 pKa = 11.84LGKK213 pKa = 7.65TLWARR218 pKa = 11.84SIGPHH223 pKa = 6.89IYY225 pKa = 10.2TIGQMSGEE233 pKa = 4.38VILRR237 pKa = 11.84DD238 pKa = 4.51GPDD241 pKa = 2.84ADD243 pKa = 3.79YY244 pKa = 11.53AVFDD248 pKa = 4.39DD249 pKa = 4.11MRR251 pKa = 11.84GGLEE255 pKa = 4.55FFHH258 pKa = 6.92GWKK261 pKa = 9.71EE262 pKa = 3.86WFGCQSVVTVKK273 pKa = 10.58KK274 pKa = 10.58LYY276 pKa = 9.92RR277 pKa = 11.84DD278 pKa = 3.77PVQMPWGKK286 pKa = 9.33PVIWLSNRR294 pKa = 11.84DD295 pKa = 3.57PRR297 pKa = 11.84DD298 pKa = 3.33EE299 pKa = 4.41LRR301 pKa = 11.84DD302 pKa = 3.78SITNHH307 pKa = 4.96TSMGKK312 pKa = 8.57QASIEE317 pKa = 4.03GDD319 pKa = 3.59IKK321 pKa = 10.77WLDD324 pKa = 3.56GNCIFVEE331 pKa = 4.06LDD333 pKa = 3.15HH334 pKa = 7.64AIFRR338 pKa = 11.84ANTEE342 pKa = 3.84
Molecular weight: 38.54 kDa
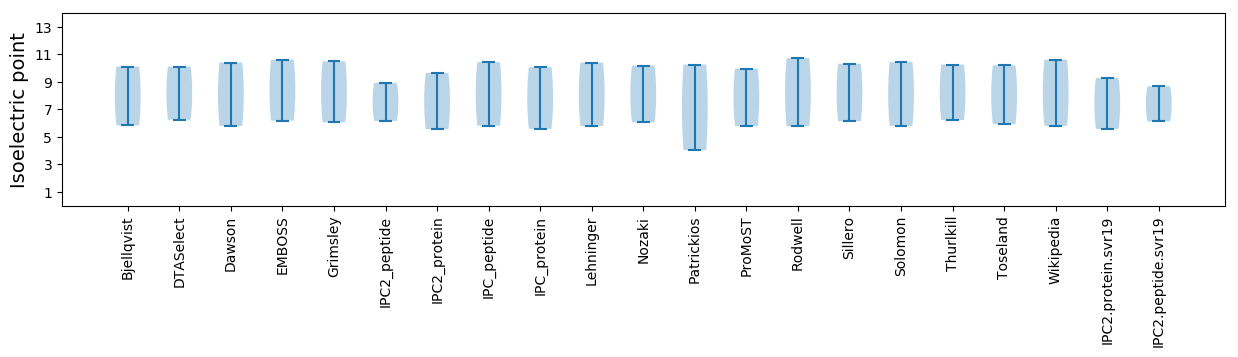
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YTC0|T1YTC0_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 3 OX=1391032 PE=4 SV=1
MM1 pKa = 7.78AYY3 pKa = 9.79RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.16SYY8 pKa = 10.45SSRR11 pKa = 11.84VKK13 pKa = 10.44RR14 pKa = 11.84RR15 pKa = 11.84TGRR18 pKa = 11.84TARR21 pKa = 11.84RR22 pKa = 11.84GGRR25 pKa = 11.84RR26 pKa = 11.84PASQQKK32 pKa = 9.42RR33 pKa = 11.84RR34 pKa = 11.84TYY36 pKa = 9.73RR37 pKa = 11.84KK38 pKa = 9.3KK39 pKa = 10.36GAMSKK44 pKa = 10.36KK45 pKa = 10.15RR46 pKa = 11.84ILNLTSRR53 pKa = 11.84KK54 pKa = 9.72KK55 pKa = 10.4RR56 pKa = 11.84DD57 pKa = 3.46TMLTWTNTTTSGGQVSPRR75 pKa = 11.84PDD77 pKa = 3.01AAYY80 pKa = 10.91VKK82 pKa = 10.26GDD84 pKa = 3.51TGGSFLWVPTARR96 pKa = 11.84DD97 pKa = 3.49LVVGSNLATIALEE110 pKa = 4.1SARR113 pKa = 11.84TSTTCYY119 pKa = 9.77MRR121 pKa = 11.84GLSEE125 pKa = 4.49KK126 pKa = 10.44LRR128 pKa = 11.84LSTSSGVPWFHH139 pKa = 7.15RR140 pKa = 11.84RR141 pKa = 11.84ICFTFKK147 pKa = 10.42GAQPFQSNFTGDD159 pKa = 3.08TGSNQLYY166 pKa = 10.44LEE168 pKa = 4.35NSNGLTRR175 pKa = 11.84LMFNQAVTIPPNYY188 pKa = 9.18QNQINSVIFKK198 pKa = 10.02GAQGVDD204 pKa = 2.93WSDD207 pKa = 3.79VIIAPIDD214 pKa = 3.39TRR216 pKa = 11.84RR217 pKa = 11.84ISLKK221 pKa = 10.16YY222 pKa = 10.23DD223 pKa = 3.26RR224 pKa = 11.84TWTYY228 pKa = 11.2RR229 pKa = 11.84SGNANGTVMEE239 pKa = 4.38KK240 pKa = 10.82KK241 pKa = 9.29MYY243 pKa = 10.66HH244 pKa = 6.38PMNKK248 pKa = 10.02NIVYY252 pKa = 10.49DD253 pKa = 4.11DD254 pKa = 4.79DD255 pKa = 4.52EE256 pKa = 6.64SGTSEE261 pKa = 3.89TTSYY265 pKa = 11.5YY266 pKa = 10.97SVDD269 pKa = 3.49SKK271 pKa = 11.82AGMGDD276 pKa = 3.78YY277 pKa = 10.33YY278 pKa = 11.29VWDD281 pKa = 4.18IFTSGPGGTATDD293 pKa = 4.87LLRR296 pKa = 11.84CDD298 pKa = 4.3ISSTLYY304 pKa = 9.11WHH306 pKa = 7.03EE307 pKa = 4.1KK308 pKa = 9.32
MM1 pKa = 7.78AYY3 pKa = 9.79RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.16SYY8 pKa = 10.45SSRR11 pKa = 11.84VKK13 pKa = 10.44RR14 pKa = 11.84RR15 pKa = 11.84TGRR18 pKa = 11.84TARR21 pKa = 11.84RR22 pKa = 11.84GGRR25 pKa = 11.84RR26 pKa = 11.84PASQQKK32 pKa = 9.42RR33 pKa = 11.84RR34 pKa = 11.84TYY36 pKa = 9.73RR37 pKa = 11.84KK38 pKa = 9.3KK39 pKa = 10.36GAMSKK44 pKa = 10.36KK45 pKa = 10.15RR46 pKa = 11.84ILNLTSRR53 pKa = 11.84KK54 pKa = 9.72KK55 pKa = 10.4RR56 pKa = 11.84DD57 pKa = 3.46TMLTWTNTTTSGGQVSPRR75 pKa = 11.84PDD77 pKa = 3.01AAYY80 pKa = 10.91VKK82 pKa = 10.26GDD84 pKa = 3.51TGGSFLWVPTARR96 pKa = 11.84DD97 pKa = 3.49LVVGSNLATIALEE110 pKa = 4.1SARR113 pKa = 11.84TSTTCYY119 pKa = 9.77MRR121 pKa = 11.84GLSEE125 pKa = 4.49KK126 pKa = 10.44LRR128 pKa = 11.84LSTSSGVPWFHH139 pKa = 7.15RR140 pKa = 11.84RR141 pKa = 11.84ICFTFKK147 pKa = 10.42GAQPFQSNFTGDD159 pKa = 3.08TGSNQLYY166 pKa = 10.44LEE168 pKa = 4.35NSNGLTRR175 pKa = 11.84LMFNQAVTIPPNYY188 pKa = 9.18QNQINSVIFKK198 pKa = 10.02GAQGVDD204 pKa = 2.93WSDD207 pKa = 3.79VIIAPIDD214 pKa = 3.39TRR216 pKa = 11.84RR217 pKa = 11.84ISLKK221 pKa = 10.16YY222 pKa = 10.23DD223 pKa = 3.26RR224 pKa = 11.84TWTYY228 pKa = 11.2RR229 pKa = 11.84SGNANGTVMEE239 pKa = 4.38KK240 pKa = 10.82KK241 pKa = 9.29MYY243 pKa = 10.66HH244 pKa = 6.38PMNKK248 pKa = 10.02NIVYY252 pKa = 10.49DD253 pKa = 4.11DD254 pKa = 4.79DD255 pKa = 4.52EE256 pKa = 6.64SGTSEE261 pKa = 3.89TTSYY265 pKa = 11.5YY266 pKa = 10.97SVDD269 pKa = 3.49SKK271 pKa = 11.82AGMGDD276 pKa = 3.78YY277 pKa = 10.33YY278 pKa = 11.29VWDD281 pKa = 4.18IFTSGPGGTATDD293 pKa = 4.87LLRR296 pKa = 11.84CDD298 pKa = 4.3ISSTLYY304 pKa = 9.11WHH306 pKa = 7.03EE307 pKa = 4.1KK308 pKa = 9.32
Molecular weight: 34.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
650 |
308 |
342 |
325.0 |
36.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.462 ± 0.592 | 1.538 ± 0.354 |
6.615 ± 0.688 | 4.154 ± 1.181 |
3.846 ± 0.58 | 9.538 ± 0.689 |
2.0 ± 0.644 | 4.923 ± 0.441 |
4.769 ± 0.879 | 6.615 ± 0.484 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.308 ± 0.386 | 3.846 ± 0.643 |
4.769 ± 0.752 | 2.462 ± 0.493 |
8.615 ± 0.503 | 7.538 ± 1.587 |
6.923 ± 2.381 | 5.692 ± 0.516 |
2.769 ± 0.312 | 4.615 ± 0.364 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
