
Dragonfly larvae associated circular virus-5
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
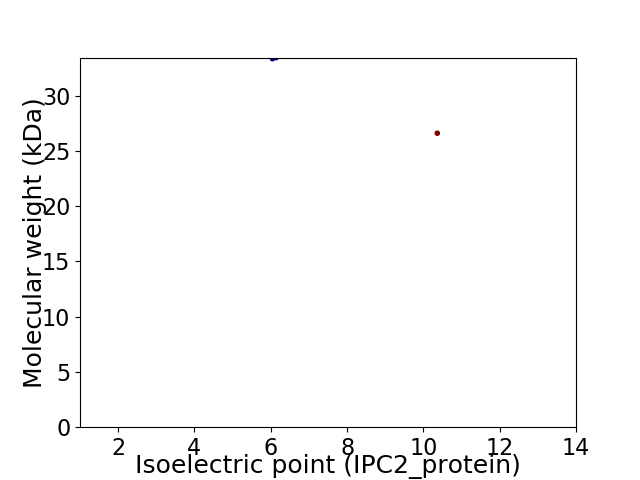
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U1V9|W5U1V9_9VIRU Coat protein OS=Dragonfly larvae associated circular virus-5 OX=1454026 PE=4 SV=1
MM1 pKa = 7.89ADD3 pKa = 4.95LLPAQQKK10 pKa = 10.34KK11 pKa = 9.45IAPTQKK17 pKa = 9.73WKK19 pKa = 10.38DD20 pKa = 3.29QRR22 pKa = 11.84GRR24 pKa = 11.84CWCFTAFVDD33 pKa = 5.97RR34 pKa = 11.84PWKK37 pKa = 10.62DD38 pKa = 3.06IACKK42 pKa = 10.27YY43 pKa = 9.7IVVGHH48 pKa = 6.61EE49 pKa = 3.83ICPKK53 pKa = 8.81TGKK56 pKa = 8.33EE57 pKa = 3.43HH58 pKa = 5.07WQGYY62 pKa = 7.35VEE64 pKa = 5.64FGTQRR69 pKa = 11.84RR70 pKa = 11.84PTGVQKK76 pKa = 9.64TLGIPDD82 pKa = 3.43LHH84 pKa = 7.57VEE86 pKa = 4.06KK87 pKa = 10.65RR88 pKa = 11.84LDD90 pKa = 3.44TSTPDD95 pKa = 3.38HH96 pKa = 6.67AANYY100 pKa = 7.99CKK102 pKa = 10.32KK103 pKa = 10.52DD104 pKa = 3.5MKK106 pKa = 10.45FVEE109 pKa = 4.2YY110 pKa = 10.78GKK112 pKa = 10.11ISKK115 pKa = 10.06QGEE118 pKa = 4.28RR119 pKa = 11.84MDD121 pKa = 5.5LDD123 pKa = 5.23LICHH127 pKa = 6.93DD128 pKa = 4.3LQTGKK133 pKa = 8.87RR134 pKa = 11.84TWRR137 pKa = 11.84EE138 pKa = 3.26IMGSDD143 pKa = 4.92DD144 pKa = 3.43VQTVCKK150 pKa = 10.46YY151 pKa = 10.99RR152 pKa = 11.84MGLQMVAAEE161 pKa = 4.37SAAEE165 pKa = 3.93RR166 pKa = 11.84AKK168 pKa = 10.65KK169 pKa = 9.35PRR171 pKa = 11.84EE172 pKa = 4.07VTWEE176 pKa = 4.19RR177 pKa = 11.84KK178 pKa = 9.42PSMAAAWLDD187 pKa = 3.7YY188 pKa = 10.82PDD190 pKa = 4.77ALMVNLNNGACWEE203 pKa = 4.25AYY205 pKa = 10.03GGEE208 pKa = 4.0EE209 pKa = 4.34EE210 pKa = 5.38IIVRR214 pKa = 11.84HH215 pKa = 5.43AKK217 pKa = 9.86SYY219 pKa = 10.48HH220 pKa = 6.47RR221 pKa = 11.84EE222 pKa = 3.85LLLDD226 pKa = 3.78GWITPLNVKK235 pKa = 8.74CTTKK239 pKa = 10.1YY240 pKa = 10.48AAWTKK245 pKa = 8.69VICVEE250 pKa = 4.27CDD252 pKa = 3.14PEE254 pKa = 4.21FFPPVEE260 pKa = 4.14PVSEE264 pKa = 4.53VVSEE268 pKa = 4.22VPPPLGNTIPGVEE281 pKa = 4.3VPAVKK286 pKa = 10.56DD287 pKa = 3.77EE288 pKa = 4.25EE289 pKa = 5.13ASALPP294 pKa = 4.0
MM1 pKa = 7.89ADD3 pKa = 4.95LLPAQQKK10 pKa = 10.34KK11 pKa = 9.45IAPTQKK17 pKa = 9.73WKK19 pKa = 10.38DD20 pKa = 3.29QRR22 pKa = 11.84GRR24 pKa = 11.84CWCFTAFVDD33 pKa = 5.97RR34 pKa = 11.84PWKK37 pKa = 10.62DD38 pKa = 3.06IACKK42 pKa = 10.27YY43 pKa = 9.7IVVGHH48 pKa = 6.61EE49 pKa = 3.83ICPKK53 pKa = 8.81TGKK56 pKa = 8.33EE57 pKa = 3.43HH58 pKa = 5.07WQGYY62 pKa = 7.35VEE64 pKa = 5.64FGTQRR69 pKa = 11.84RR70 pKa = 11.84PTGVQKK76 pKa = 9.64TLGIPDD82 pKa = 3.43LHH84 pKa = 7.57VEE86 pKa = 4.06KK87 pKa = 10.65RR88 pKa = 11.84LDD90 pKa = 3.44TSTPDD95 pKa = 3.38HH96 pKa = 6.67AANYY100 pKa = 7.99CKK102 pKa = 10.32KK103 pKa = 10.52DD104 pKa = 3.5MKK106 pKa = 10.45FVEE109 pKa = 4.2YY110 pKa = 10.78GKK112 pKa = 10.11ISKK115 pKa = 10.06QGEE118 pKa = 4.28RR119 pKa = 11.84MDD121 pKa = 5.5LDD123 pKa = 5.23LICHH127 pKa = 6.93DD128 pKa = 4.3LQTGKK133 pKa = 8.87RR134 pKa = 11.84TWRR137 pKa = 11.84EE138 pKa = 3.26IMGSDD143 pKa = 4.92DD144 pKa = 3.43VQTVCKK150 pKa = 10.46YY151 pKa = 10.99RR152 pKa = 11.84MGLQMVAAEE161 pKa = 4.37SAAEE165 pKa = 3.93RR166 pKa = 11.84AKK168 pKa = 10.65KK169 pKa = 9.35PRR171 pKa = 11.84EE172 pKa = 4.07VTWEE176 pKa = 4.19RR177 pKa = 11.84KK178 pKa = 9.42PSMAAAWLDD187 pKa = 3.7YY188 pKa = 10.82PDD190 pKa = 4.77ALMVNLNNGACWEE203 pKa = 4.25AYY205 pKa = 10.03GGEE208 pKa = 4.0EE209 pKa = 4.34EE210 pKa = 5.38IIVRR214 pKa = 11.84HH215 pKa = 5.43AKK217 pKa = 9.86SYY219 pKa = 10.48HH220 pKa = 6.47RR221 pKa = 11.84EE222 pKa = 3.85LLLDD226 pKa = 3.78GWITPLNVKK235 pKa = 8.74CTTKK239 pKa = 10.1YY240 pKa = 10.48AAWTKK245 pKa = 8.69VICVEE250 pKa = 4.27CDD252 pKa = 3.14PEE254 pKa = 4.21FFPPVEE260 pKa = 4.14PVSEE264 pKa = 4.53VVSEE268 pKa = 4.22VPPPLGNTIPGVEE281 pKa = 4.3VPAVKK286 pKa = 10.56DD287 pKa = 3.77EE288 pKa = 4.25EE289 pKa = 5.13ASALPP294 pKa = 4.0
Molecular weight: 33.35 kDa
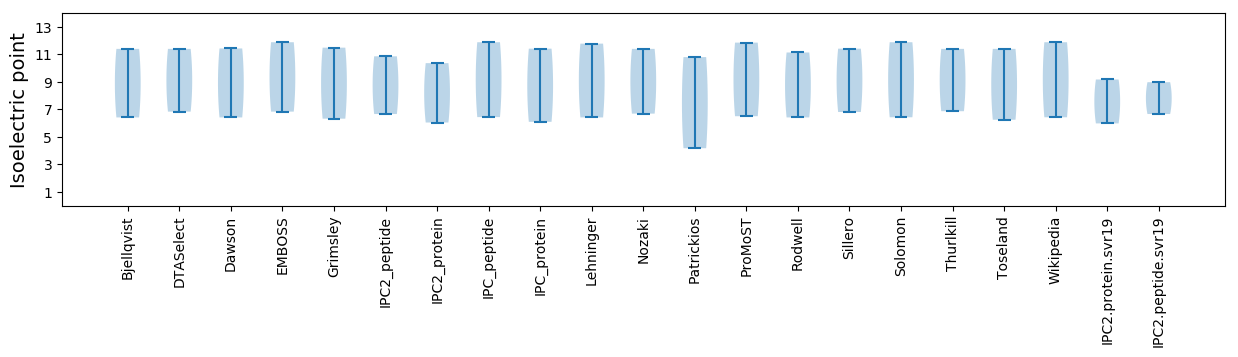
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U1V9|W5U1V9_9VIRU Coat protein OS=Dragonfly larvae associated circular virus-5 OX=1454026 PE=4 SV=1
MM1 pKa = 6.89VFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84VSRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84ASRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84VRR19 pKa = 11.84RR20 pKa = 11.84TSRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84PRR27 pKa = 11.84ARR29 pKa = 11.84RR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84NGGNSIMSSSIRR45 pKa = 11.84MPHH48 pKa = 6.7KK49 pKa = 9.42YY50 pKa = 10.35SEE52 pKa = 4.26IFGLTFTGGSGVAGIYY68 pKa = 9.75QFYY71 pKa = 10.73LNYY74 pKa = 10.15LEE76 pKa = 4.96HH77 pKa = 7.11PGYY80 pKa = 9.3TGAGHH85 pKa = 6.04QPMGFDD91 pKa = 4.06QISALFNRR99 pKa = 11.84YY100 pKa = 8.15RR101 pKa = 11.84VIGMSYY107 pKa = 10.38HH108 pKa = 6.49IALTNISGVYY118 pKa = 7.26VAEE121 pKa = 3.9YY122 pKa = 10.27AVYY125 pKa = 10.0YY126 pKa = 9.17RR127 pKa = 11.84PNMTVTNDD135 pKa = 2.9FTAIMEE141 pKa = 4.47GTGVLAKK148 pKa = 10.07GHH150 pKa = 6.63LGITSSGSDD159 pKa = 2.98TRR161 pKa = 11.84THH163 pKa = 6.59RR164 pKa = 11.84GYY166 pKa = 11.35VSVARR171 pKa = 11.84ARR173 pKa = 11.84GVPRR177 pKa = 11.84RR178 pKa = 11.84NVMTEE183 pKa = 3.24SDD185 pKa = 3.82YY186 pKa = 11.38QAILGVAPNFLPSFNIAVVNQDD208 pKa = 2.71ASASVTLRR216 pKa = 11.84VRR218 pKa = 11.84VDD220 pKa = 3.12LKK222 pKa = 11.1FYY224 pKa = 9.06TIWSDD229 pKa = 3.36RR230 pKa = 11.84KK231 pKa = 10.96VMGTSS236 pKa = 2.98
MM1 pKa = 6.89VFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84VSRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84ASRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84VRR19 pKa = 11.84RR20 pKa = 11.84TSRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84PRR27 pKa = 11.84ARR29 pKa = 11.84RR30 pKa = 11.84GRR32 pKa = 11.84RR33 pKa = 11.84NGGNSIMSSSIRR45 pKa = 11.84MPHH48 pKa = 6.7KK49 pKa = 9.42YY50 pKa = 10.35SEE52 pKa = 4.26IFGLTFTGGSGVAGIYY68 pKa = 9.75QFYY71 pKa = 10.73LNYY74 pKa = 10.15LEE76 pKa = 4.96HH77 pKa = 7.11PGYY80 pKa = 9.3TGAGHH85 pKa = 6.04QPMGFDD91 pKa = 4.06QISALFNRR99 pKa = 11.84YY100 pKa = 8.15RR101 pKa = 11.84VIGMSYY107 pKa = 10.38HH108 pKa = 6.49IALTNISGVYY118 pKa = 7.26VAEE121 pKa = 3.9YY122 pKa = 10.27AVYY125 pKa = 10.0YY126 pKa = 9.17RR127 pKa = 11.84PNMTVTNDD135 pKa = 2.9FTAIMEE141 pKa = 4.47GTGVLAKK148 pKa = 10.07GHH150 pKa = 6.63LGITSSGSDD159 pKa = 2.98TRR161 pKa = 11.84THH163 pKa = 6.59RR164 pKa = 11.84GYY166 pKa = 11.35VSVARR171 pKa = 11.84ARR173 pKa = 11.84GVPRR177 pKa = 11.84RR178 pKa = 11.84NVMTEE183 pKa = 3.24SDD185 pKa = 3.82YY186 pKa = 11.38QAILGVAPNFLPSFNIAVVNQDD208 pKa = 2.71ASASVTLRR216 pKa = 11.84VRR218 pKa = 11.84VDD220 pKa = 3.12LKK222 pKa = 11.1FYY224 pKa = 9.06TIWSDD229 pKa = 3.36RR230 pKa = 11.84KK231 pKa = 10.96VMGTSS236 pKa = 2.98
Molecular weight: 26.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
530 |
236 |
294 |
265.0 |
29.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.736 ± 0.351 | 2.075 ± 1.367 |
4.717 ± 1.153 | 5.472 ± 2.208 |
3.019 ± 0.802 | 7.736 ± 1.324 |
2.453 ± 0.059 | 4.906 ± 0.397 |
5.472 ± 2.487 | 5.472 ± 0.534 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.208 ± 0.399 | 3.208 ± 0.957 |
5.472 ± 1.371 | 3.019 ± 0.593 |
9.057 ± 3.245 | 5.849 ± 2.287 |
6.226 ± 0.364 | 8.491 ± 0.269 |
2.075 ± 1.088 | 4.34 ± 1.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
