
White clover mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
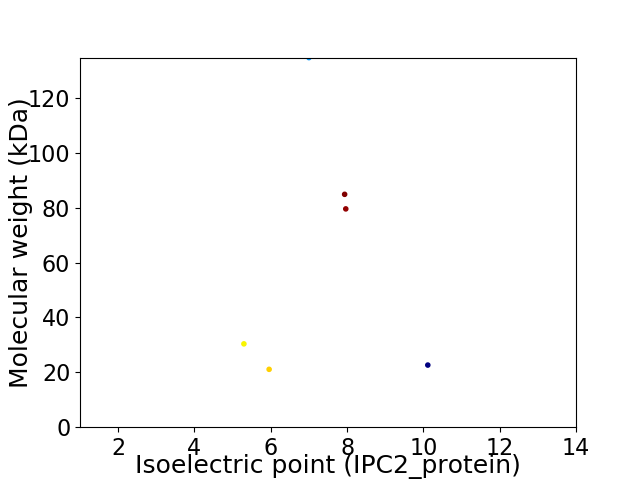
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1J1DTY0|A0A1J1DTY0_9LUTE Serine protease OS=White clover mottle virus OX=1913024 PE=4 SV=1
MM1 pKa = 7.41NFLINNQNSRR11 pKa = 11.84IEE13 pKa = 4.14LTFSNSLSLDD23 pKa = 3.27TRR25 pKa = 11.84RR26 pKa = 11.84SLTALFLISAEE37 pKa = 4.01QYY39 pKa = 10.4LQISHH44 pKa = 6.06EE45 pKa = 4.02QHH47 pKa = 5.34GTQNSVAYY55 pKa = 10.09CSLLVCSFSTRR66 pKa = 11.84GIQFPRR72 pKa = 11.84WQTCHH77 pKa = 6.61PLEE80 pKa = 4.54FEE82 pKa = 4.18KK83 pKa = 11.05SPPLRR88 pKa = 11.84ALPRR92 pKa = 11.84AFGEE96 pKa = 4.69GEE98 pKa = 4.0FCILSSSEE106 pKa = 3.57NGAAEE111 pKa = 4.64LYY113 pKa = 10.6SPNPNSCHH121 pKa = 6.36RR122 pKa = 11.84ANLYY126 pKa = 10.4RR127 pKa = 11.84LTYY130 pKa = 10.39SSLAEE135 pKa = 3.93GLPRR139 pKa = 11.84YY140 pKa = 9.58PEE142 pKa = 3.64IFGYY146 pKa = 8.14GTSFIQTLIGSYY158 pKa = 9.13IRR160 pKa = 11.84KK161 pKa = 8.97IEE163 pKa = 3.97RR164 pKa = 11.84GVFFLRR170 pKa = 11.84PVFPMGDD177 pKa = 3.29SLGVDD182 pKa = 3.65LRR184 pKa = 11.84TLGFLLLDD192 pKa = 3.28GRR194 pKa = 11.84GYY196 pKa = 10.63NNQYY200 pKa = 8.91HH201 pKa = 7.29AYY203 pKa = 7.89YY204 pKa = 9.97SSRR207 pKa = 11.84IANVLHH213 pKa = 7.03RR214 pKa = 11.84IYY216 pKa = 10.95GKK218 pKa = 10.49SFTLAFWRR226 pKa = 11.84LLQLDD231 pKa = 4.99CGPCFQVPSLPMEE244 pKa = 4.13APIAEE249 pKa = 4.64GTCGQDD255 pKa = 3.57SDD257 pKa = 6.07DD258 pKa = 4.42EE259 pKa = 5.76GEE261 pKa = 4.23DD262 pKa = 4.65DD263 pKa = 3.95EE264 pKa = 6.23GVWEE268 pKa = 4.21LL269 pKa = 4.43
MM1 pKa = 7.41NFLINNQNSRR11 pKa = 11.84IEE13 pKa = 4.14LTFSNSLSLDD23 pKa = 3.27TRR25 pKa = 11.84RR26 pKa = 11.84SLTALFLISAEE37 pKa = 4.01QYY39 pKa = 10.4LQISHH44 pKa = 6.06EE45 pKa = 4.02QHH47 pKa = 5.34GTQNSVAYY55 pKa = 10.09CSLLVCSFSTRR66 pKa = 11.84GIQFPRR72 pKa = 11.84WQTCHH77 pKa = 6.61PLEE80 pKa = 4.54FEE82 pKa = 4.18KK83 pKa = 11.05SPPLRR88 pKa = 11.84ALPRR92 pKa = 11.84AFGEE96 pKa = 4.69GEE98 pKa = 4.0FCILSSSEE106 pKa = 3.57NGAAEE111 pKa = 4.64LYY113 pKa = 10.6SPNPNSCHH121 pKa = 6.36RR122 pKa = 11.84ANLYY126 pKa = 10.4RR127 pKa = 11.84LTYY130 pKa = 10.39SSLAEE135 pKa = 3.93GLPRR139 pKa = 11.84YY140 pKa = 9.58PEE142 pKa = 3.64IFGYY146 pKa = 8.14GTSFIQTLIGSYY158 pKa = 9.13IRR160 pKa = 11.84KK161 pKa = 8.97IEE163 pKa = 3.97RR164 pKa = 11.84GVFFLRR170 pKa = 11.84PVFPMGDD177 pKa = 3.29SLGVDD182 pKa = 3.65LRR184 pKa = 11.84TLGFLLLDD192 pKa = 3.28GRR194 pKa = 11.84GYY196 pKa = 10.63NNQYY200 pKa = 8.91HH201 pKa = 7.29AYY203 pKa = 7.89YY204 pKa = 9.97SSRR207 pKa = 11.84IANVLHH213 pKa = 7.03RR214 pKa = 11.84IYY216 pKa = 10.95GKK218 pKa = 10.49SFTLAFWRR226 pKa = 11.84LLQLDD231 pKa = 4.99CGPCFQVPSLPMEE244 pKa = 4.13APIAEE249 pKa = 4.64GTCGQDD255 pKa = 3.57SDD257 pKa = 6.07DD258 pKa = 4.42EE259 pKa = 5.76GEE261 pKa = 4.23DD262 pKa = 4.65DD263 pKa = 3.95EE264 pKa = 6.23GVWEE268 pKa = 4.21LL269 pKa = 4.43
Molecular weight: 30.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1J1DP29|A0A1J1DP29_9LUTE p0 protein OS=White clover mottle virus OX=1913024 PE=4 SV=1
MM1 pKa = 6.53NTVVVRR7 pKa = 11.84NNGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84PIRR20 pKa = 11.84RR21 pKa = 11.84AQRR24 pKa = 11.84RR25 pKa = 11.84NPVVVVQPARR35 pKa = 11.84QPQRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84AARR52 pKa = 11.84GSTAGRR58 pKa = 11.84RR59 pKa = 11.84GSSEE63 pKa = 3.79TFIFSKK69 pKa = 11.13DD70 pKa = 3.4NLKK73 pKa = 11.05GSDD76 pKa = 3.11SGRR79 pKa = 11.84ITFGPSLSDD88 pKa = 3.49CPAFSSGILKK98 pKa = 10.26AYY100 pKa = 10.13HH101 pKa = 6.54EE102 pKa = 4.61YY103 pKa = 10.53KK104 pKa = 9.31ITMVKK109 pKa = 10.64LEE111 pKa = 5.07FISEE115 pKa = 4.18AASTSSGSIAYY126 pKa = 9.43EE127 pKa = 3.84LDD129 pKa = 3.51PHH131 pKa = 6.89CKK133 pKa = 9.94ASEE136 pKa = 3.93LGSYY140 pKa = 9.5INKK143 pKa = 10.01FGITKK148 pKa = 10.23SGQRR152 pKa = 11.84TFSARR157 pKa = 11.84FINGIEE163 pKa = 3.94WHH165 pKa = 6.88SSDD168 pKa = 3.83EE169 pKa = 4.08DD170 pKa = 3.46QFRR173 pKa = 11.84ILYY176 pKa = 9.46KK177 pKa = 11.06GNGNSAIAGSFRR189 pKa = 11.84ITIKK193 pKa = 10.84CQTQNAKK200 pKa = 10.37
MM1 pKa = 6.53NTVVVRR7 pKa = 11.84NNGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84PIRR20 pKa = 11.84RR21 pKa = 11.84AQRR24 pKa = 11.84RR25 pKa = 11.84NPVVVVQPARR35 pKa = 11.84QPQRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84AARR52 pKa = 11.84GSTAGRR58 pKa = 11.84RR59 pKa = 11.84GSSEE63 pKa = 3.79TFIFSKK69 pKa = 11.13DD70 pKa = 3.4NLKK73 pKa = 11.05GSDD76 pKa = 3.11SGRR79 pKa = 11.84ITFGPSLSDD88 pKa = 3.49CPAFSSGILKK98 pKa = 10.26AYY100 pKa = 10.13HH101 pKa = 6.54EE102 pKa = 4.61YY103 pKa = 10.53KK104 pKa = 9.31ITMVKK109 pKa = 10.64LEE111 pKa = 5.07FISEE115 pKa = 4.18AASTSSGSIAYY126 pKa = 9.43EE127 pKa = 3.84LDD129 pKa = 3.51PHH131 pKa = 6.89CKK133 pKa = 9.94ASEE136 pKa = 3.93LGSYY140 pKa = 9.5INKK143 pKa = 10.01FGITKK148 pKa = 10.23SGQRR152 pKa = 11.84TFSARR157 pKa = 11.84FINGIEE163 pKa = 3.94WHH165 pKa = 6.88SSDD168 pKa = 3.83EE169 pKa = 4.08DD170 pKa = 3.46QFRR173 pKa = 11.84ILYY176 pKa = 9.46KK177 pKa = 11.06GNGNSAIAGSFRR189 pKa = 11.84ITIKK193 pKa = 10.84CQTQNAKK200 pKa = 10.37
Molecular weight: 22.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3296 |
193 |
1178 |
549.3 |
62.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.916 ± 0.193 | 1.881 ± 0.23 |
4.733 ± 0.396 | 6.796 ± 0.325 |
3.975 ± 0.308 | 6.493 ± 0.362 |
2.154 ± 0.37 | 5.036 ± 0.262 |
6.614 ± 0.81 | 8.677 ± 1.127 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.002 ± 0.216 | 5.036 ± 0.276 |
5.431 ± 0.408 | 3.914 ± 0.209 |
6.22 ± 1.104 | 8.799 ± 0.616 |
5.067 ± 0.499 | 5.674 ± 0.532 |
2.124 ± 0.251 | 3.428 ± 0.212 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
