
Spider monkey simian foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Simiispumavirus
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
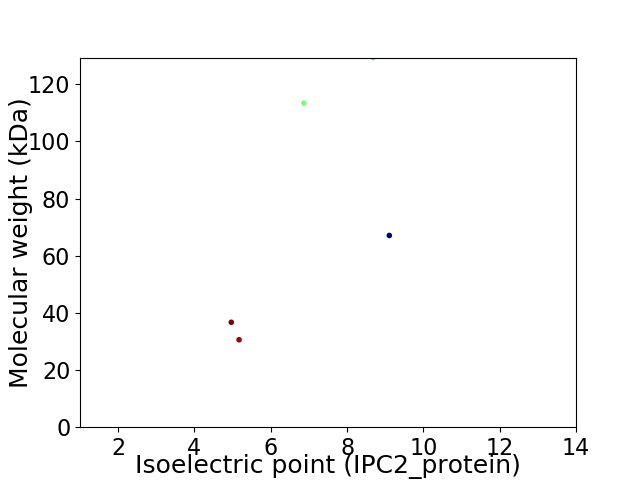
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8HC81|A8HC81_9RETR Bel2 OS=Spider monkey simian foamy virus OX=2170200 GN=bel2 PE=4 SV=1
MM1 pKa = 8.15ASQQEE6 pKa = 4.43KK7 pKa = 10.78NSIVPEE13 pKa = 4.24DD14 pKa = 3.95DD15 pKa = 3.54FMEE18 pKa = 4.37EE19 pKa = 3.54LRR21 pKa = 11.84AFLAEE26 pKa = 4.37FPIAEE31 pKa = 4.38QPLEE35 pKa = 4.41DD36 pKa = 5.61DD37 pKa = 3.88VFQAEE42 pKa = 4.74GQQPSQFLEE51 pKa = 4.48DD52 pKa = 4.06PGEE55 pKa = 4.57GPSGASAQAGASSGPPKK72 pKa = 10.73VSGKK76 pKa = 9.0LWNMMAHH83 pKa = 6.44HH84 pKa = 7.42PNRR87 pKa = 11.84GTTSHH92 pKa = 6.64GPDD95 pKa = 3.42EE96 pKa = 4.97PPTGRR101 pKa = 11.84EE102 pKa = 3.42PRR104 pKa = 11.84LRR106 pKa = 11.84WLQEE110 pKa = 3.26ARR112 pKa = 11.84MEE114 pKa = 3.93TAYY117 pKa = 9.87IRR119 pKa = 11.84FEE121 pKa = 3.97KK122 pKa = 10.61DD123 pKa = 2.78LEE125 pKa = 4.5KK126 pKa = 10.84FFPTPEE132 pKa = 3.61QRR134 pKa = 11.84EE135 pKa = 4.04FFIQRR140 pKa = 11.84AIITAAGYY148 pKa = 10.43KK149 pKa = 9.45PDD151 pKa = 3.6LAIYY155 pKa = 7.3TAHH158 pKa = 6.51NGWYY162 pKa = 10.38ACIQPHH168 pKa = 5.94TGDD171 pKa = 3.59LGEE174 pKa = 4.63KK175 pKa = 9.65YY176 pKa = 9.56EE177 pKa = 4.62VYY179 pKa = 10.5YY180 pKa = 10.98KK181 pKa = 10.57CLKK184 pKa = 10.09CGNEE188 pKa = 3.63QWDD191 pKa = 3.91PLLYY195 pKa = 10.68DD196 pKa = 3.91WDD198 pKa = 4.3PSVLMFLRR206 pKa = 11.84AWWKK210 pKa = 8.46TPHH213 pKa = 6.31TSSPLTAMRR222 pKa = 11.84RR223 pKa = 11.84HH224 pKa = 6.44DD225 pKa = 3.95AVCAGIYY232 pKa = 9.82SDD234 pKa = 4.52SSSSTEE240 pKa = 3.66PPKK243 pKa = 10.36RR244 pKa = 11.84RR245 pKa = 11.84QRR247 pKa = 11.84GDD249 pKa = 3.15PGHH252 pKa = 6.75RR253 pKa = 11.84WGAYY257 pKa = 8.88KK258 pKa = 9.59RR259 pKa = 11.84TFDD262 pKa = 4.85SGPLEE267 pKa = 4.04QLVLTHH273 pKa = 6.2TCAYY277 pKa = 5.07TTEE280 pKa = 4.0WPEE283 pKa = 3.76PKK285 pKa = 9.65RR286 pKa = 11.84IHH288 pKa = 6.82LGDD291 pKa = 4.46LEE293 pKa = 4.87TVCPALPDD301 pKa = 4.27PGEE304 pKa = 4.46GGTNSEE310 pKa = 5.07MEE312 pKa = 4.01PQYY315 pKa = 11.09QQGSWTFDD323 pKa = 3.03QQ324 pKa = 4.84
MM1 pKa = 8.15ASQQEE6 pKa = 4.43KK7 pKa = 10.78NSIVPEE13 pKa = 4.24DD14 pKa = 3.95DD15 pKa = 3.54FMEE18 pKa = 4.37EE19 pKa = 3.54LRR21 pKa = 11.84AFLAEE26 pKa = 4.37FPIAEE31 pKa = 4.38QPLEE35 pKa = 4.41DD36 pKa = 5.61DD37 pKa = 3.88VFQAEE42 pKa = 4.74GQQPSQFLEE51 pKa = 4.48DD52 pKa = 4.06PGEE55 pKa = 4.57GPSGASAQAGASSGPPKK72 pKa = 10.73VSGKK76 pKa = 9.0LWNMMAHH83 pKa = 6.44HH84 pKa = 7.42PNRR87 pKa = 11.84GTTSHH92 pKa = 6.64GPDD95 pKa = 3.42EE96 pKa = 4.97PPTGRR101 pKa = 11.84EE102 pKa = 3.42PRR104 pKa = 11.84LRR106 pKa = 11.84WLQEE110 pKa = 3.26ARR112 pKa = 11.84MEE114 pKa = 3.93TAYY117 pKa = 9.87IRR119 pKa = 11.84FEE121 pKa = 3.97KK122 pKa = 10.61DD123 pKa = 2.78LEE125 pKa = 4.5KK126 pKa = 10.84FFPTPEE132 pKa = 3.61QRR134 pKa = 11.84EE135 pKa = 4.04FFIQRR140 pKa = 11.84AIITAAGYY148 pKa = 10.43KK149 pKa = 9.45PDD151 pKa = 3.6LAIYY155 pKa = 7.3TAHH158 pKa = 6.51NGWYY162 pKa = 10.38ACIQPHH168 pKa = 5.94TGDD171 pKa = 3.59LGEE174 pKa = 4.63KK175 pKa = 9.65YY176 pKa = 9.56EE177 pKa = 4.62VYY179 pKa = 10.5YY180 pKa = 10.98KK181 pKa = 10.57CLKK184 pKa = 10.09CGNEE188 pKa = 3.63QWDD191 pKa = 3.91PLLYY195 pKa = 10.68DD196 pKa = 3.91WDD198 pKa = 4.3PSVLMFLRR206 pKa = 11.84AWWKK210 pKa = 8.46TPHH213 pKa = 6.31TSSPLTAMRR222 pKa = 11.84RR223 pKa = 11.84HH224 pKa = 6.44DD225 pKa = 3.95AVCAGIYY232 pKa = 9.82SDD234 pKa = 4.52SSSSTEE240 pKa = 3.66PPKK243 pKa = 10.36RR244 pKa = 11.84RR245 pKa = 11.84QRR247 pKa = 11.84GDD249 pKa = 3.15PGHH252 pKa = 6.75RR253 pKa = 11.84WGAYY257 pKa = 8.88KK258 pKa = 9.59RR259 pKa = 11.84TFDD262 pKa = 4.85SGPLEE267 pKa = 4.04QLVLTHH273 pKa = 6.2TCAYY277 pKa = 5.07TTEE280 pKa = 4.0WPEE283 pKa = 3.76PKK285 pKa = 9.65RR286 pKa = 11.84IHH288 pKa = 6.82LGDD291 pKa = 4.46LEE293 pKa = 4.87TVCPALPDD301 pKa = 4.27PGEE304 pKa = 4.46GGTNSEE310 pKa = 5.07MEE312 pKa = 4.01PQYY315 pKa = 11.09QQGSWTFDD323 pKa = 3.03QQ324 pKa = 4.84
Molecular weight: 36.71 kDa
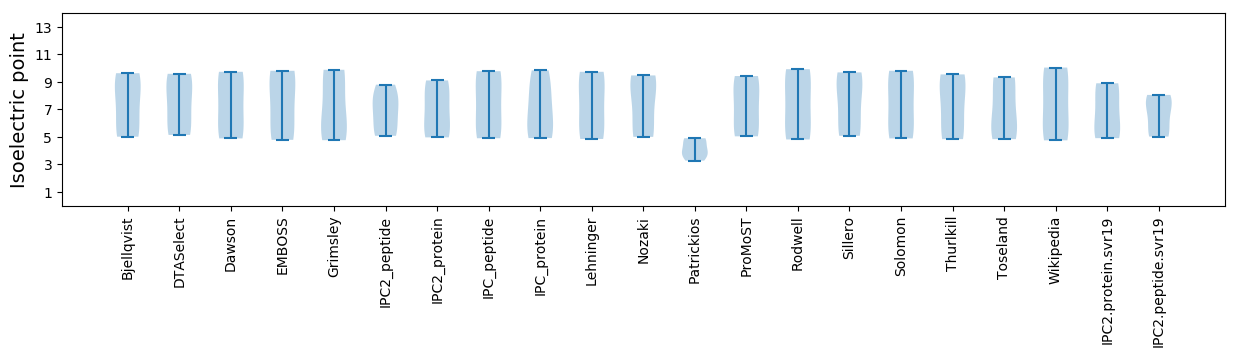
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8HC78|A8HC78_9RETR Pr125Pol OS=Spider monkey simian foamy virus OX=2170200 GN=pol PE=4 SV=1
MM1 pKa = 7.33AQGNQPEE8 pKa = 4.94HH9 pKa = 7.77IDD11 pKa = 3.55IGHH14 pKa = 6.22LTILLRR20 pKa = 11.84EE21 pKa = 4.38NGLPTNPPHH30 pKa = 5.13GTEE33 pKa = 3.86YY34 pKa = 10.71AVRR37 pKa = 11.84MTEE40 pKa = 4.31GWWGDD45 pKa = 3.46YY46 pKa = 11.19ARR48 pKa = 11.84FTVIRR53 pKa = 11.84VAFQDD58 pKa = 3.6NQGNPLPRR66 pKa = 11.84PEE68 pKa = 3.87WEE70 pKa = 4.2YY71 pKa = 10.47INRR74 pKa = 11.84DD75 pKa = 3.33VRR77 pKa = 11.84PLNDD81 pKa = 4.26SIIGCTFRR89 pKa = 11.84QAQQAFNNIDD99 pKa = 3.77LARR102 pKa = 11.84TPSRR106 pKa = 11.84YY107 pKa = 10.02GPLSNGLFLITDD119 pKa = 4.29PEE121 pKa = 4.28WLTFNPLSAVEE132 pKa = 4.63IADD135 pKa = 5.39LDD137 pKa = 3.87DD138 pKa = 3.94QEE140 pKa = 5.27IRR142 pKa = 11.84DD143 pKa = 4.3FLAHH147 pKa = 5.87ATIVLDD153 pKa = 3.61NVMADD158 pKa = 3.19VVIQQRR164 pKa = 11.84EE165 pKa = 4.15IEE167 pKa = 4.94DD168 pKa = 3.47LTKK171 pKa = 10.83TNLRR175 pKa = 11.84LRR177 pKa = 11.84NALASSGTYY186 pKa = 9.48QPIGMPIAQSSPAGPSASVAGPHH209 pKa = 5.64MPAMPSVPYY218 pKa = 10.48ISAIPSAPPAPIPVAPAAAPPLGPLPSAPAPPVLPVIQPNTFGTDD263 pKa = 3.24GNGYY267 pKa = 7.18TAVGPVPVLPIAQIKK282 pKa = 9.55AVIGEE287 pKa = 4.34APTDD291 pKa = 3.44ARR293 pKa = 11.84QIPLWVAKK301 pKa = 9.76HH302 pKa = 5.27AAAIEE307 pKa = 4.52GTFPTGSADD316 pKa = 3.29VRR318 pKa = 11.84CRR320 pKa = 11.84VLNALLTSHH329 pKa = 6.74GGMTLSPNEE338 pKa = 4.21CGTWSLAAAALYY350 pKa = 9.59QRR352 pKa = 11.84IYY354 pKa = 11.23GVIPLHH360 pKa = 6.64DD361 pKa = 5.1LPHH364 pKa = 6.12TMAEE368 pKa = 4.07VARR371 pKa = 11.84RR372 pKa = 11.84EE373 pKa = 4.41GILVAFNMGMTFTNNSFDD391 pKa = 4.06VVWGIIRR398 pKa = 11.84PLLPGQASVAMLQGYY413 pKa = 9.51LDD415 pKa = 3.76QYY417 pKa = 11.18QGQQQKK423 pKa = 10.29AQAFPTLLRR432 pKa = 11.84RR433 pKa = 11.84TFEE436 pKa = 4.37TLGLNYY442 pKa = 9.94LGQSIRR448 pKa = 11.84SNQPARR454 pKa = 11.84TSGSVAGSITQGRR467 pKa = 11.84GRR469 pKa = 11.84GQPRR473 pKa = 11.84SRR475 pKa = 11.84PPNRR479 pKa = 11.84ASNRR483 pKa = 11.84SNNPPRR489 pKa = 11.84PPPPRR494 pKa = 11.84SQEE497 pKa = 4.07QQPSPAGRR505 pKa = 11.84GSSSSSAPPQRR516 pKa = 11.84SGYY519 pKa = 8.17NLRR522 pKa = 11.84QQINRR527 pKa = 11.84PQRR530 pKa = 11.84LGGGPNPYY538 pKa = 10.18RR539 pKa = 11.84NLEE542 pKa = 3.82NRR544 pKa = 11.84NIPRR548 pKa = 11.84SEE550 pKa = 4.08TVPSSRR556 pKa = 11.84RR557 pKa = 11.84AEE559 pKa = 3.93TSEE562 pKa = 4.02TNRR565 pKa = 11.84PNQARR570 pKa = 11.84GGQQGSSNRR579 pKa = 11.84NQTRR583 pKa = 11.84SINTVRR589 pKa = 11.84ASDD592 pKa = 3.75SQVIPSAPPAEE603 pKa = 4.35TNPAATAVTPNGPNQHH619 pKa = 6.39SGRR622 pKa = 11.84NN623 pKa = 3.57
MM1 pKa = 7.33AQGNQPEE8 pKa = 4.94HH9 pKa = 7.77IDD11 pKa = 3.55IGHH14 pKa = 6.22LTILLRR20 pKa = 11.84EE21 pKa = 4.38NGLPTNPPHH30 pKa = 5.13GTEE33 pKa = 3.86YY34 pKa = 10.71AVRR37 pKa = 11.84MTEE40 pKa = 4.31GWWGDD45 pKa = 3.46YY46 pKa = 11.19ARR48 pKa = 11.84FTVIRR53 pKa = 11.84VAFQDD58 pKa = 3.6NQGNPLPRR66 pKa = 11.84PEE68 pKa = 3.87WEE70 pKa = 4.2YY71 pKa = 10.47INRR74 pKa = 11.84DD75 pKa = 3.33VRR77 pKa = 11.84PLNDD81 pKa = 4.26SIIGCTFRR89 pKa = 11.84QAQQAFNNIDD99 pKa = 3.77LARR102 pKa = 11.84TPSRR106 pKa = 11.84YY107 pKa = 10.02GPLSNGLFLITDD119 pKa = 4.29PEE121 pKa = 4.28WLTFNPLSAVEE132 pKa = 4.63IADD135 pKa = 5.39LDD137 pKa = 3.87DD138 pKa = 3.94QEE140 pKa = 5.27IRR142 pKa = 11.84DD143 pKa = 4.3FLAHH147 pKa = 5.87ATIVLDD153 pKa = 3.61NVMADD158 pKa = 3.19VVIQQRR164 pKa = 11.84EE165 pKa = 4.15IEE167 pKa = 4.94DD168 pKa = 3.47LTKK171 pKa = 10.83TNLRR175 pKa = 11.84LRR177 pKa = 11.84NALASSGTYY186 pKa = 9.48QPIGMPIAQSSPAGPSASVAGPHH209 pKa = 5.64MPAMPSVPYY218 pKa = 10.48ISAIPSAPPAPIPVAPAAAPPLGPLPSAPAPPVLPVIQPNTFGTDD263 pKa = 3.24GNGYY267 pKa = 7.18TAVGPVPVLPIAQIKK282 pKa = 9.55AVIGEE287 pKa = 4.34APTDD291 pKa = 3.44ARR293 pKa = 11.84QIPLWVAKK301 pKa = 9.76HH302 pKa = 5.27AAAIEE307 pKa = 4.52GTFPTGSADD316 pKa = 3.29VRR318 pKa = 11.84CRR320 pKa = 11.84VLNALLTSHH329 pKa = 6.74GGMTLSPNEE338 pKa = 4.21CGTWSLAAAALYY350 pKa = 9.59QRR352 pKa = 11.84IYY354 pKa = 11.23GVIPLHH360 pKa = 6.64DD361 pKa = 5.1LPHH364 pKa = 6.12TMAEE368 pKa = 4.07VARR371 pKa = 11.84RR372 pKa = 11.84EE373 pKa = 4.41GILVAFNMGMTFTNNSFDD391 pKa = 4.06VVWGIIRR398 pKa = 11.84PLLPGQASVAMLQGYY413 pKa = 9.51LDD415 pKa = 3.76QYY417 pKa = 11.18QGQQQKK423 pKa = 10.29AQAFPTLLRR432 pKa = 11.84RR433 pKa = 11.84TFEE436 pKa = 4.37TLGLNYY442 pKa = 9.94LGQSIRR448 pKa = 11.84SNQPARR454 pKa = 11.84TSGSVAGSITQGRR467 pKa = 11.84GRR469 pKa = 11.84GQPRR473 pKa = 11.84SRR475 pKa = 11.84PPNRR479 pKa = 11.84ASNRR483 pKa = 11.84SNNPPRR489 pKa = 11.84PPPPRR494 pKa = 11.84SQEE497 pKa = 4.07QQPSPAGRR505 pKa = 11.84GSSSSSAPPQRR516 pKa = 11.84SGYY519 pKa = 8.17NLRR522 pKa = 11.84QQINRR527 pKa = 11.84PQRR530 pKa = 11.84LGGGPNPYY538 pKa = 10.18RR539 pKa = 11.84NLEE542 pKa = 3.82NRR544 pKa = 11.84NIPRR548 pKa = 11.84SEE550 pKa = 4.08TVPSSRR556 pKa = 11.84RR557 pKa = 11.84AEE559 pKa = 3.93TSEE562 pKa = 4.02TNRR565 pKa = 11.84PNQARR570 pKa = 11.84GGQQGSSNRR579 pKa = 11.84NQTRR583 pKa = 11.84SINTVRR589 pKa = 11.84ASDD592 pKa = 3.75SQVIPSAPPAEE603 pKa = 4.35TNPAATAVTPNGPNQHH619 pKa = 6.39SGRR622 pKa = 11.84NN623 pKa = 3.57
Molecular weight: 67.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3349 |
268 |
1148 |
669.8 |
75.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.271 ± 0.943 | 1.583 ± 0.699 |
4.598 ± 0.405 | 5.524 ± 0.777 |
2.717 ± 0.262 | 5.673 ± 0.698 |
2.448 ± 0.242 | 6.659 ± 0.645 |
5.464 ± 1.248 | 9.645 ± 0.81 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.732 ± 0.256 | 5.046 ± 0.668 |
7.943 ± 1.037 | 5.345 ± 0.32 |
4.897 ± 0.727 | 6.42 ± 0.445 |
7.077 ± 0.344 | 5.255 ± 0.537 |
2.21 ± 0.346 | 3.494 ± 0.366 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
