
[Collinsella] massiliensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Enorma
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
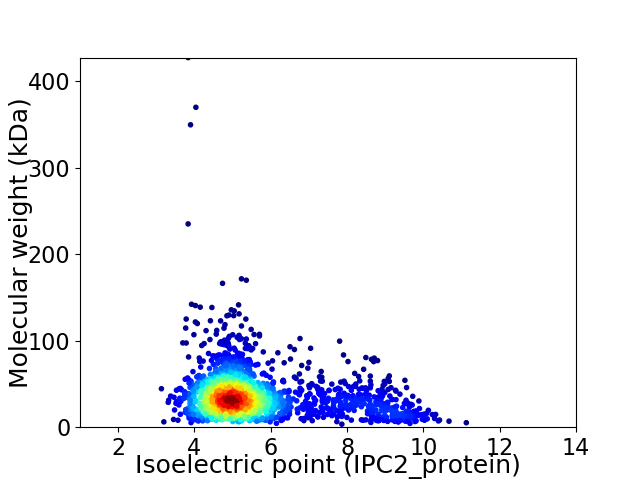
Virtual 2D-PAGE plot for 1896 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
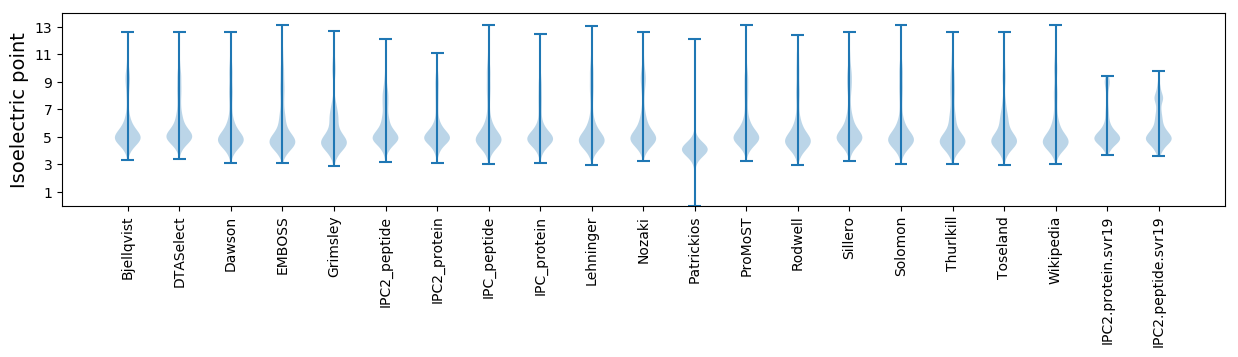
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y3XZ00|A0A1Y3XZ00_9ACTN Uncharacterized protein OS=[Collinsella] massiliensis OX=1232426 GN=B5G02_06680 PE=4 SV=1
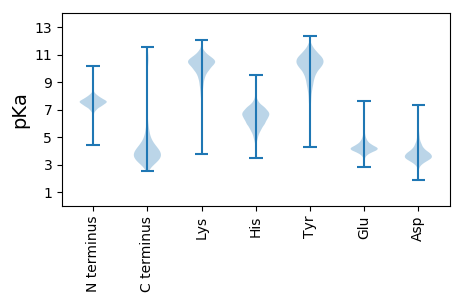
MM1 pKa = 6.6YY2 pKa = 10.09QPSISRR8 pKa = 11.84RR9 pKa = 11.84GLIATLAAASALPLAACGSEE29 pKa = 3.99DD30 pKa = 3.47AKK32 pKa = 11.0EE33 pKa = 4.24GSSTGKK39 pKa = 10.58ASASDD44 pKa = 3.01GKK46 pKa = 8.9TYY48 pKa = 10.58KK49 pKa = 10.26IGVLQLTQHH58 pKa = 6.84AALDD62 pKa = 3.68QTNEE66 pKa = 3.9GFIKK70 pKa = 10.76ALDD73 pKa = 3.98EE74 pKa = 5.03AGLSYY79 pKa = 11.16DD80 pKa = 4.79ADD82 pKa = 3.67QQNASNDD89 pKa = 3.32QTACQTIAQTFVNDD103 pKa = 3.79GSDD106 pKa = 3.79LIFAIGTPAAQAVAAATKK124 pKa = 8.64EE125 pKa = 3.91IPIVGSAITDD135 pKa = 4.17FEE137 pKa = 4.88SVGLVSSNDD146 pKa = 3.36APGGNVTGSSDD157 pKa = 3.75LTPVSDD163 pKa = 5.32QIDD166 pKa = 4.33LLVKK170 pKa = 10.0LVPDD174 pKa = 4.62CKK176 pKa = 10.19TAGILYY182 pKa = 7.7CTAEE186 pKa = 4.46SNSTIQVDD194 pKa = 3.94LAKK197 pKa = 10.28EE198 pKa = 3.83ALEE201 pKa = 5.1SYY203 pKa = 10.44DD204 pKa = 3.75LPYY207 pKa = 11.3EE208 pKa = 4.13EE209 pKa = 5.07FTVSSSNEE217 pKa = 3.62IQSVVEE223 pKa = 3.98TAVGKK228 pKa = 10.57VDD230 pKa = 5.84AIYY233 pKa = 10.77APTDD237 pKa = 3.38NTIAAAMAQVSSIADD252 pKa = 3.51EE253 pKa = 4.64AGVPVIVGCDD263 pKa = 3.28TMVEE267 pKa = 4.21DD268 pKa = 5.55GGTASYY274 pKa = 10.36SINYY278 pKa = 8.85VDD280 pKa = 4.96LGHH283 pKa = 7.19KK284 pKa = 9.68AGEE287 pKa = 3.98MAVRR291 pKa = 11.84ILTEE295 pKa = 4.16GEE297 pKa = 4.16NPADD301 pKa = 3.75MPIEE305 pKa = 3.94YY306 pKa = 10.01LDD308 pKa = 4.33ASEE311 pKa = 4.73CTLIANQASADD322 pKa = 3.61ACGVDD327 pKa = 5.94LGILDD332 pKa = 3.64NPKK335 pKa = 9.85IVV337 pKa = 3.15
MM1 pKa = 6.6YY2 pKa = 10.09QPSISRR8 pKa = 11.84RR9 pKa = 11.84GLIATLAAASALPLAACGSEE29 pKa = 3.99DD30 pKa = 3.47AKK32 pKa = 11.0EE33 pKa = 4.24GSSTGKK39 pKa = 10.58ASASDD44 pKa = 3.01GKK46 pKa = 8.9TYY48 pKa = 10.58KK49 pKa = 10.26IGVLQLTQHH58 pKa = 6.84AALDD62 pKa = 3.68QTNEE66 pKa = 3.9GFIKK70 pKa = 10.76ALDD73 pKa = 3.98EE74 pKa = 5.03AGLSYY79 pKa = 11.16DD80 pKa = 4.79ADD82 pKa = 3.67QQNASNDD89 pKa = 3.32QTACQTIAQTFVNDD103 pKa = 3.79GSDD106 pKa = 3.79LIFAIGTPAAQAVAAATKK124 pKa = 8.64EE125 pKa = 3.91IPIVGSAITDD135 pKa = 4.17FEE137 pKa = 4.88SVGLVSSNDD146 pKa = 3.36APGGNVTGSSDD157 pKa = 3.75LTPVSDD163 pKa = 5.32QIDD166 pKa = 4.33LLVKK170 pKa = 10.0LVPDD174 pKa = 4.62CKK176 pKa = 10.19TAGILYY182 pKa = 7.7CTAEE186 pKa = 4.46SNSTIQVDD194 pKa = 3.94LAKK197 pKa = 10.28EE198 pKa = 3.83ALEE201 pKa = 5.1SYY203 pKa = 10.44DD204 pKa = 3.75LPYY207 pKa = 11.3EE208 pKa = 4.13EE209 pKa = 5.07FTVSSSNEE217 pKa = 3.62IQSVVEE223 pKa = 3.98TAVGKK228 pKa = 10.57VDD230 pKa = 5.84AIYY233 pKa = 10.77APTDD237 pKa = 3.38NTIAAAMAQVSSIADD252 pKa = 3.51EE253 pKa = 4.64AGVPVIVGCDD263 pKa = 3.28TMVEE267 pKa = 4.21DD268 pKa = 5.55GGTASYY274 pKa = 10.36SINYY278 pKa = 8.85VDD280 pKa = 4.96LGHH283 pKa = 7.19KK284 pKa = 9.68AGEE287 pKa = 3.98MAVRR291 pKa = 11.84ILTEE295 pKa = 4.16GEE297 pKa = 4.16NPADD301 pKa = 3.75MPIEE305 pKa = 3.94YY306 pKa = 10.01LDD308 pKa = 4.33ASEE311 pKa = 4.73CTLIANQASADD322 pKa = 3.61ACGVDD327 pKa = 5.94LGILDD332 pKa = 3.64NPKK335 pKa = 9.85IVV337 pKa = 3.15
Molecular weight: 34.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y3XW37|A0A1Y3XW37_9ACTN Transcription termination factor Rho OS=[Collinsella] massiliensis OX=1232426 GN=rho PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.62RR10 pKa = 11.84KK11 pKa = 9.56RR12 pKa = 11.84AKK14 pKa = 8.48THH16 pKa = 5.36GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.37GGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.34QPNKK9 pKa = 8.62RR10 pKa = 11.84KK11 pKa = 9.56RR12 pKa = 11.84AKK14 pKa = 8.48THH16 pKa = 5.36GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.37GGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
653081 |
31 |
3985 |
344.5 |
37.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.496 ± 0.083 | 1.482 ± 0.026 |
6.389 ± 0.055 | 6.846 ± 0.052 |
3.608 ± 0.033 | 8.256 ± 0.049 |
1.919 ± 0.027 | 5.186 ± 0.049 |
3.058 ± 0.054 | 9.171 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.59 ± 0.028 | 2.582 ± 0.043 |
4.489 ± 0.036 | 2.777 ± 0.031 |
6.558 ± 0.082 | 5.396 ± 0.043 |
5.44 ± 0.088 | 7.903 ± 0.049 |
1.016 ± 0.021 | 2.84 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
