
Rhodobacteraceae bacterium KMS-5
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; unclassified Rhodobacteraceae
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
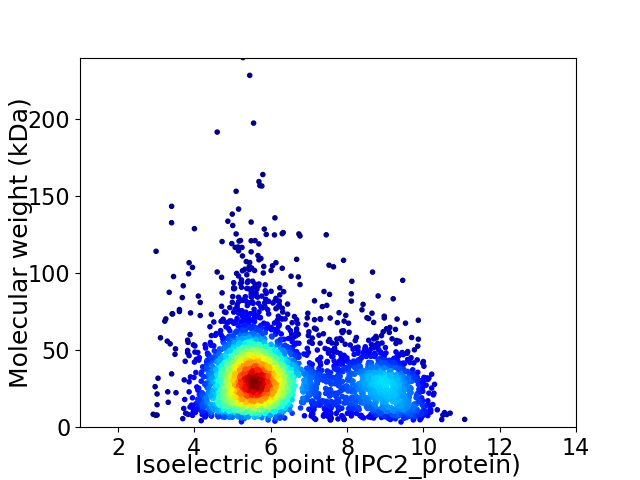
Virtual 2D-PAGE plot for 3633 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M0QU51|A0A6M0QU51_9RHOB HutD family protein OS=Rhodobacteraceae bacterium KMS-5 OX=2710650 GN=G4Z14_07655 PE=4 SV=1
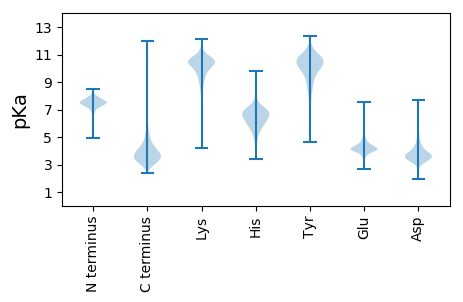
MM1 pKa = 7.86ADD3 pKa = 4.29DD4 pKa = 5.16IYY6 pKa = 11.04TIDD9 pKa = 3.65ALATGSEE16 pKa = 4.74TITITDD22 pKa = 4.65DD23 pKa = 3.53GTGSDD28 pKa = 3.08WLVYY32 pKa = 9.51SGSYY36 pKa = 10.51ASYY39 pKa = 11.62AEE41 pKa = 4.87IDD43 pKa = 3.91LSWFVDD49 pKa = 3.48SGISQSAEE57 pKa = 3.25GLYY60 pKa = 8.83FTVTGGHH67 pKa = 6.02RR68 pKa = 11.84LVINGLIEE76 pKa = 4.01NARR79 pKa = 11.84GTTAEE84 pKa = 4.22DD85 pKa = 3.84WISGNEE91 pKa = 3.96ADD93 pKa = 4.11NLLYY97 pKa = 10.98GDD99 pKa = 5.04FDD101 pKa = 3.8ATGAGGNDD109 pKa = 4.49TINGDD114 pKa = 3.49AGRR117 pKa = 11.84DD118 pKa = 3.67TIYY121 pKa = 10.89GGSGLDD127 pKa = 3.66SLEE130 pKa = 3.95GSYY133 pKa = 11.51DD134 pKa = 3.85DD135 pKa = 6.2DD136 pKa = 4.92LLHH139 pKa = 7.29GDD141 pKa = 4.12GGADD145 pKa = 3.37TVSGGAGNDD154 pKa = 3.39TVLGGEE160 pKa = 4.38GADD163 pKa = 3.39WLYY166 pKa = 11.57GGADD170 pKa = 3.28ARR172 pKa = 11.84DD173 pKa = 3.71VVSYY177 pKa = 11.34ADD179 pKa = 3.64STDD182 pKa = 2.97GVYY185 pKa = 10.04IEE187 pKa = 4.36ITYY190 pKa = 10.76GSTTIATSGFAEE202 pKa = 4.4GDD204 pKa = 3.67HH205 pKa = 7.03LNGFTDD211 pKa = 5.62AIGSAYY217 pKa = 10.07GDD219 pKa = 4.34RR220 pKa = 11.84ITDD223 pKa = 3.67TVVNTIAFGYY233 pKa = 8.42NANRR237 pKa = 11.84FYY239 pKa = 11.38GGGGADD245 pKa = 4.42LLNMGGGNDD254 pKa = 3.39SGYY257 pKa = 11.18GGDD260 pKa = 4.92GRR262 pKa = 11.84DD263 pKa = 3.5SVNGGVGDD271 pKa = 4.29DD272 pKa = 4.18LLFGGGGNDD281 pKa = 4.47DD282 pKa = 3.56VDD284 pKa = 4.21GGTGADD290 pKa = 3.14VVYY293 pKa = 10.81GGDD296 pKa = 3.52GYY298 pKa = 11.79NRR300 pKa = 11.84LIGGAGQDD308 pKa = 3.28RR309 pKa = 11.84VIGGGLRR316 pKa = 11.84EE317 pKa = 4.08TLYY320 pKa = 11.24GGAGDD325 pKa = 4.19DD326 pKa = 3.69TLLAGGGNDD335 pKa = 4.33LLFANEE341 pKa = 3.94GNDD344 pKa = 3.27RR345 pKa = 11.84VLGGAGDD352 pKa = 4.15DD353 pKa = 4.41LLFGLAGKK361 pKa = 8.18DD362 pKa = 3.46TLIGGAGADD371 pKa = 3.31EE372 pKa = 4.49FRR374 pKa = 11.84FISATDD380 pKa = 3.59GGSDD384 pKa = 3.07AASRR388 pKa = 11.84DD389 pKa = 3.29VVMDD393 pKa = 4.17FNRR396 pKa = 11.84SQGDD400 pKa = 3.54HH401 pKa = 6.37LVFSIAAASLDD412 pKa = 4.05FIGQQSFTGQAGEE425 pKa = 3.89VRR427 pKa = 11.84FIVTAQGTRR436 pKa = 11.84AFADD440 pKa = 3.46LDD442 pKa = 3.59GDD444 pKa = 4.82RR445 pKa = 11.84ITDD448 pKa = 3.29QAFDD452 pKa = 3.63VLGVSLLQEE461 pKa = 4.04SDD463 pKa = 3.75FVLL466 pKa = 4.49
MM1 pKa = 7.86ADD3 pKa = 4.29DD4 pKa = 5.16IYY6 pKa = 11.04TIDD9 pKa = 3.65ALATGSEE16 pKa = 4.74TITITDD22 pKa = 4.65DD23 pKa = 3.53GTGSDD28 pKa = 3.08WLVYY32 pKa = 9.51SGSYY36 pKa = 10.51ASYY39 pKa = 11.62AEE41 pKa = 4.87IDD43 pKa = 3.91LSWFVDD49 pKa = 3.48SGISQSAEE57 pKa = 3.25GLYY60 pKa = 8.83FTVTGGHH67 pKa = 6.02RR68 pKa = 11.84LVINGLIEE76 pKa = 4.01NARR79 pKa = 11.84GTTAEE84 pKa = 4.22DD85 pKa = 3.84WISGNEE91 pKa = 3.96ADD93 pKa = 4.11NLLYY97 pKa = 10.98GDD99 pKa = 5.04FDD101 pKa = 3.8ATGAGGNDD109 pKa = 4.49TINGDD114 pKa = 3.49AGRR117 pKa = 11.84DD118 pKa = 3.67TIYY121 pKa = 10.89GGSGLDD127 pKa = 3.66SLEE130 pKa = 3.95GSYY133 pKa = 11.51DD134 pKa = 3.85DD135 pKa = 6.2DD136 pKa = 4.92LLHH139 pKa = 7.29GDD141 pKa = 4.12GGADD145 pKa = 3.37TVSGGAGNDD154 pKa = 3.39TVLGGEE160 pKa = 4.38GADD163 pKa = 3.39WLYY166 pKa = 11.57GGADD170 pKa = 3.28ARR172 pKa = 11.84DD173 pKa = 3.71VVSYY177 pKa = 11.34ADD179 pKa = 3.64STDD182 pKa = 2.97GVYY185 pKa = 10.04IEE187 pKa = 4.36ITYY190 pKa = 10.76GSTTIATSGFAEE202 pKa = 4.4GDD204 pKa = 3.67HH205 pKa = 7.03LNGFTDD211 pKa = 5.62AIGSAYY217 pKa = 10.07GDD219 pKa = 4.34RR220 pKa = 11.84ITDD223 pKa = 3.67TVVNTIAFGYY233 pKa = 8.42NANRR237 pKa = 11.84FYY239 pKa = 11.38GGGGADD245 pKa = 4.42LLNMGGGNDD254 pKa = 3.39SGYY257 pKa = 11.18GGDD260 pKa = 4.92GRR262 pKa = 11.84DD263 pKa = 3.5SVNGGVGDD271 pKa = 4.29DD272 pKa = 4.18LLFGGGGNDD281 pKa = 4.47DD282 pKa = 3.56VDD284 pKa = 4.21GGTGADD290 pKa = 3.14VVYY293 pKa = 10.81GGDD296 pKa = 3.52GYY298 pKa = 11.79NRR300 pKa = 11.84LIGGAGQDD308 pKa = 3.28RR309 pKa = 11.84VIGGGLRR316 pKa = 11.84EE317 pKa = 4.08TLYY320 pKa = 11.24GGAGDD325 pKa = 4.19DD326 pKa = 3.69TLLAGGGNDD335 pKa = 4.33LLFANEE341 pKa = 3.94GNDD344 pKa = 3.27RR345 pKa = 11.84VLGGAGDD352 pKa = 4.15DD353 pKa = 4.41LLFGLAGKK361 pKa = 8.18DD362 pKa = 3.46TLIGGAGADD371 pKa = 3.31EE372 pKa = 4.49FRR374 pKa = 11.84FISATDD380 pKa = 3.59GGSDD384 pKa = 3.07AASRR388 pKa = 11.84DD389 pKa = 3.29VVMDD393 pKa = 4.17FNRR396 pKa = 11.84SQGDD400 pKa = 3.54HH401 pKa = 6.37LVFSIAAASLDD412 pKa = 4.05FIGQQSFTGQAGEE425 pKa = 3.89VRR427 pKa = 11.84FIVTAQGTRR436 pKa = 11.84AFADD440 pKa = 3.46LDD442 pKa = 3.59GDD444 pKa = 4.82RR445 pKa = 11.84ITDD448 pKa = 3.29QAFDD452 pKa = 3.63VLGVSLLQEE461 pKa = 4.04SDD463 pKa = 3.75FVLL466 pKa = 4.49
Molecular weight: 47.37 kDa
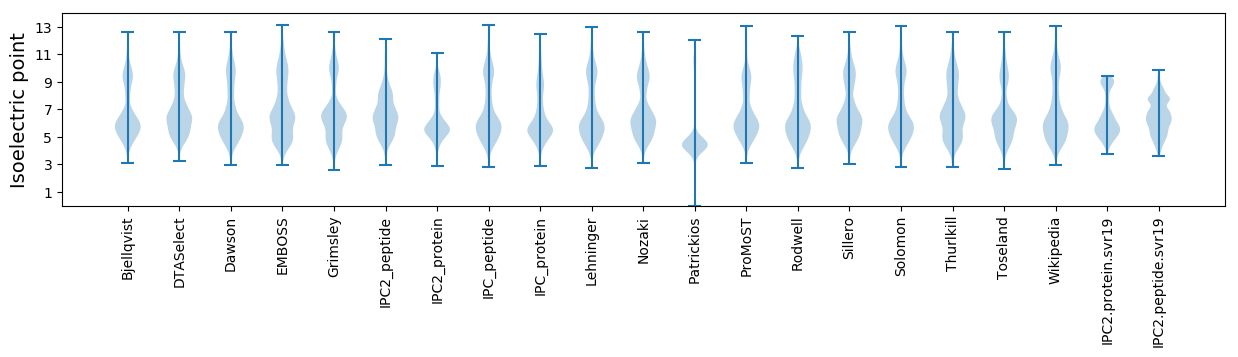
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M0QYF1|A0A6M0QYF1_9RHOB Lytic transglycosylase domain-containing protein OS=Rhodobacteraceae bacterium KMS-5 OX=2710650 GN=G4Z14_17155 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.21GRR39 pKa = 11.84HH40 pKa = 5.51KK41 pKa = 10.9LSAA44 pKa = 3.8
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34GGRR28 pKa = 11.84LVLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.21GRR39 pKa = 11.84HH40 pKa = 5.51KK41 pKa = 10.9LSAA44 pKa = 3.8
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1152034 |
30 |
2141 |
317.1 |
34.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.654 ± 0.067 | 0.872 ± 0.013 |
5.663 ± 0.035 | 5.59 ± 0.037 |
3.623 ± 0.027 | 9.174 ± 0.051 |
2.094 ± 0.021 | 4.765 ± 0.029 |
3.069 ± 0.034 | 10.432 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.706 ± 0.02 | 2.375 ± 0.025 |
5.25 ± 0.034 | 3.051 ± 0.021 |
6.88 ± 0.041 | 4.704 ± 0.028 |
5.289 ± 0.031 | 7.294 ± 0.034 |
1.441 ± 0.02 | 2.076 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
