
Gemmiger sp. An120
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Gemmiger; unclassified Gemmiger
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
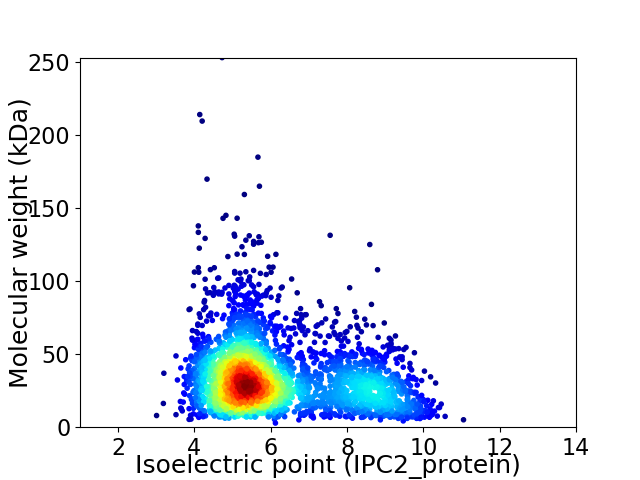
Virtual 2D-PAGE plot for 3048 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4TL65|A0A1Y4TL65_9FIRM Peptide ABC transporter permease OS=Gemmiger sp. An120 OX=1965549 GN=B5E65_09290 PE=3 SV=1
MM1 pKa = 7.3NRR3 pKa = 11.84TDD5 pKa = 3.31IIKK8 pKa = 10.53RR9 pKa = 11.84IAAGVLAAVLTAALAGCGKK28 pKa = 8.44TPASSVGGQDD38 pKa = 2.83TFEE41 pKa = 5.14PDD43 pKa = 3.27SAVSTTGAWRR53 pKa = 11.84EE54 pKa = 4.03EE55 pKa = 4.24EE56 pKa = 4.23VTPDD60 pKa = 3.03EE61 pKa = 4.29NARR64 pKa = 11.84SYY66 pKa = 8.17KK67 pKa = 10.29TPVTLADD74 pKa = 3.53GSVRR78 pKa = 11.84MLEE81 pKa = 4.06AGADD85 pKa = 3.77NTLHH89 pKa = 7.12LLTSADD95 pKa = 5.03NGATWQEE102 pKa = 4.11ITVDD106 pKa = 3.55WQPAGAEE113 pKa = 4.23SFGVCRR119 pKa = 11.84MLPDD123 pKa = 4.17GGCFVTAVKK132 pKa = 10.3EE133 pKa = 4.21DD134 pKa = 3.02NGQRR138 pKa = 11.84KK139 pKa = 8.19EE140 pKa = 4.37SYY142 pKa = 7.35WTLDD146 pKa = 3.25PEE148 pKa = 4.66TGVLDD153 pKa = 4.13PVTLPEE159 pKa = 5.08NIVQLSDD166 pKa = 3.62AYY168 pKa = 10.07PLGGDD173 pKa = 3.66SLLLLGMSLVPASADD188 pKa = 3.3APAVLVDD195 pKa = 3.98EE196 pKa = 4.74NGMMKK201 pKa = 8.06TTSPWKK207 pKa = 10.44LDD209 pKa = 3.76LATGACEE216 pKa = 4.4GLTALTNTLQGSIISGMAADD236 pKa = 3.96PTEE239 pKa = 4.08LGAYY243 pKa = 9.78YY244 pKa = 10.61SLAYY248 pKa = 9.5MDD250 pKa = 4.26TGVQLLRR257 pKa = 11.84TDD259 pKa = 3.62ADD261 pKa = 3.6EE262 pKa = 4.41ATACVFDD269 pKa = 4.31SLPNASAAAFAACADD284 pKa = 3.7AEE286 pKa = 4.42GNYY289 pKa = 10.38YY290 pKa = 9.83YY291 pKa = 10.98ASSKK295 pKa = 10.55GIYY298 pKa = 9.26RR299 pKa = 11.84VAKK302 pKa = 10.47GGDD305 pKa = 3.36LAEE308 pKa = 4.43LVVDD312 pKa = 4.92GASTALSRR320 pKa = 11.84SEE322 pKa = 3.95AVVMGLTCCADD333 pKa = 3.31GSFLTVSLEE342 pKa = 4.06GGQDD346 pKa = 2.86AMTAHH351 pKa = 8.4LYY353 pKa = 9.78RR354 pKa = 11.84YY355 pKa = 9.24YY356 pKa = 10.39WDD358 pKa = 3.97ADD360 pKa = 3.5APAEE364 pKa = 4.08EE365 pKa = 4.51STAALTVWSLEE376 pKa = 4.06NNDD379 pKa = 3.46TVRR382 pKa = 11.84AAIEE386 pKa = 4.23VYY388 pKa = 9.51EE389 pKa = 4.01QQNPGLTVTYY399 pKa = 9.94QVAVEE404 pKa = 4.71DD405 pKa = 4.37GATTRR410 pKa = 11.84EE411 pKa = 4.17DD412 pKa = 3.58ALSALNTALLAGSGPDD428 pKa = 3.61VLILDD433 pKa = 4.11GLDD436 pKa = 2.76WQAYY440 pKa = 7.83RR441 pKa = 11.84DD442 pKa = 3.85KK443 pKa = 11.51GLLADD448 pKa = 4.2LSEE451 pKa = 4.23WVDD454 pKa = 4.38LSALQSNLVDD464 pKa = 3.59PFRR467 pKa = 11.84EE468 pKa = 4.0QEE470 pKa = 4.34GVFVLPARR478 pKa = 11.84FSVPVLCGAPEE489 pKa = 4.22DD490 pKa = 4.18LAGLDD495 pKa = 3.91SLDD498 pKa = 3.89ALAEE502 pKa = 4.06YY503 pKa = 10.72LLQLPARR510 pKa = 11.84PAWDD514 pKa = 3.62YY515 pKa = 11.31TSAGYY520 pKa = 10.1YY521 pKa = 8.88EE522 pKa = 4.53QLEE525 pKa = 4.32PPYY528 pKa = 10.68GLGFVSVEE536 pKa = 3.77QLLDD540 pKa = 3.79FVLGSSAAALTEE552 pKa = 4.6DD553 pKa = 4.41GANGDD558 pKa = 3.5AVEE561 pKa = 4.37AVLKK565 pKa = 9.76FVQQVGRR572 pKa = 11.84YY573 pKa = 9.67YY574 pKa = 11.44GMEE577 pKa = 4.27DD578 pKa = 3.5YY579 pKa = 10.88KK580 pKa = 11.49GLLSNGVISSGSEE593 pKa = 4.05GQSVSYY599 pKa = 11.36GDD601 pKa = 3.76GSYY604 pKa = 11.16EE605 pKa = 4.37CFSTGNALLAWEE617 pKa = 4.41EE618 pKa = 4.17MLTPSYY624 pKa = 9.61LTAVRR629 pKa = 11.84KK630 pKa = 9.98DD631 pKa = 3.31RR632 pKa = 11.84AGVSLIQRR640 pKa = 11.84PGLVQGAYY648 pKa = 8.55TPRR651 pKa = 11.84TLAAVCADD659 pKa = 3.18ASEE662 pKa = 4.12EE663 pKa = 3.92AGAFVAILFGSEE675 pKa = 4.28VQASWQQDD683 pKa = 3.63GMPVRR688 pKa = 11.84ADD690 pKa = 3.37ALQSSIEE697 pKa = 4.18ANCPDD702 pKa = 3.67PEE704 pKa = 4.44ARR706 pKa = 11.84QNVQALIDD714 pKa = 3.99ALQTPVTQPDD724 pKa = 3.57DD725 pKa = 3.8TVRR728 pKa = 11.84DD729 pKa = 3.71ALLQNANALIAGDD742 pKa = 3.67VTLEE746 pKa = 3.81QARR749 pKa = 11.84TGVEE753 pKa = 3.96DD754 pKa = 4.41ALNLYY759 pKa = 9.86LAEE762 pKa = 4.19QEE764 pKa = 4.22
MM1 pKa = 7.3NRR3 pKa = 11.84TDD5 pKa = 3.31IIKK8 pKa = 10.53RR9 pKa = 11.84IAAGVLAAVLTAALAGCGKK28 pKa = 8.44TPASSVGGQDD38 pKa = 2.83TFEE41 pKa = 5.14PDD43 pKa = 3.27SAVSTTGAWRR53 pKa = 11.84EE54 pKa = 4.03EE55 pKa = 4.24EE56 pKa = 4.23VTPDD60 pKa = 3.03EE61 pKa = 4.29NARR64 pKa = 11.84SYY66 pKa = 8.17KK67 pKa = 10.29TPVTLADD74 pKa = 3.53GSVRR78 pKa = 11.84MLEE81 pKa = 4.06AGADD85 pKa = 3.77NTLHH89 pKa = 7.12LLTSADD95 pKa = 5.03NGATWQEE102 pKa = 4.11ITVDD106 pKa = 3.55WQPAGAEE113 pKa = 4.23SFGVCRR119 pKa = 11.84MLPDD123 pKa = 4.17GGCFVTAVKK132 pKa = 10.3EE133 pKa = 4.21DD134 pKa = 3.02NGQRR138 pKa = 11.84KK139 pKa = 8.19EE140 pKa = 4.37SYY142 pKa = 7.35WTLDD146 pKa = 3.25PEE148 pKa = 4.66TGVLDD153 pKa = 4.13PVTLPEE159 pKa = 5.08NIVQLSDD166 pKa = 3.62AYY168 pKa = 10.07PLGGDD173 pKa = 3.66SLLLLGMSLVPASADD188 pKa = 3.3APAVLVDD195 pKa = 3.98EE196 pKa = 4.74NGMMKK201 pKa = 8.06TTSPWKK207 pKa = 10.44LDD209 pKa = 3.76LATGACEE216 pKa = 4.4GLTALTNTLQGSIISGMAADD236 pKa = 3.96PTEE239 pKa = 4.08LGAYY243 pKa = 9.78YY244 pKa = 10.61SLAYY248 pKa = 9.5MDD250 pKa = 4.26TGVQLLRR257 pKa = 11.84TDD259 pKa = 3.62ADD261 pKa = 3.6EE262 pKa = 4.41ATACVFDD269 pKa = 4.31SLPNASAAAFAACADD284 pKa = 3.7AEE286 pKa = 4.42GNYY289 pKa = 10.38YY290 pKa = 9.83YY291 pKa = 10.98ASSKK295 pKa = 10.55GIYY298 pKa = 9.26RR299 pKa = 11.84VAKK302 pKa = 10.47GGDD305 pKa = 3.36LAEE308 pKa = 4.43LVVDD312 pKa = 4.92GASTALSRR320 pKa = 11.84SEE322 pKa = 3.95AVVMGLTCCADD333 pKa = 3.31GSFLTVSLEE342 pKa = 4.06GGQDD346 pKa = 2.86AMTAHH351 pKa = 8.4LYY353 pKa = 9.78RR354 pKa = 11.84YY355 pKa = 9.24YY356 pKa = 10.39WDD358 pKa = 3.97ADD360 pKa = 3.5APAEE364 pKa = 4.08EE365 pKa = 4.51STAALTVWSLEE376 pKa = 4.06NNDD379 pKa = 3.46TVRR382 pKa = 11.84AAIEE386 pKa = 4.23VYY388 pKa = 9.51EE389 pKa = 4.01QQNPGLTVTYY399 pKa = 9.94QVAVEE404 pKa = 4.71DD405 pKa = 4.37GATTRR410 pKa = 11.84EE411 pKa = 4.17DD412 pKa = 3.58ALSALNTALLAGSGPDD428 pKa = 3.61VLILDD433 pKa = 4.11GLDD436 pKa = 2.76WQAYY440 pKa = 7.83RR441 pKa = 11.84DD442 pKa = 3.85KK443 pKa = 11.51GLLADD448 pKa = 4.2LSEE451 pKa = 4.23WVDD454 pKa = 4.38LSALQSNLVDD464 pKa = 3.59PFRR467 pKa = 11.84EE468 pKa = 4.0QEE470 pKa = 4.34GVFVLPARR478 pKa = 11.84FSVPVLCGAPEE489 pKa = 4.22DD490 pKa = 4.18LAGLDD495 pKa = 3.91SLDD498 pKa = 3.89ALAEE502 pKa = 4.06YY503 pKa = 10.72LLQLPARR510 pKa = 11.84PAWDD514 pKa = 3.62YY515 pKa = 11.31TSAGYY520 pKa = 10.1YY521 pKa = 8.88EE522 pKa = 4.53QLEE525 pKa = 4.32PPYY528 pKa = 10.68GLGFVSVEE536 pKa = 3.77QLLDD540 pKa = 3.79FVLGSSAAALTEE552 pKa = 4.6DD553 pKa = 4.41GANGDD558 pKa = 3.5AVEE561 pKa = 4.37AVLKK565 pKa = 9.76FVQQVGRR572 pKa = 11.84YY573 pKa = 9.67YY574 pKa = 11.44GMEE577 pKa = 4.27DD578 pKa = 3.5YY579 pKa = 10.88KK580 pKa = 11.49GLLSNGVISSGSEE593 pKa = 4.05GQSVSYY599 pKa = 11.36GDD601 pKa = 3.76GSYY604 pKa = 11.16EE605 pKa = 4.37CFSTGNALLAWEE617 pKa = 4.41EE618 pKa = 4.17MLTPSYY624 pKa = 9.61LTAVRR629 pKa = 11.84KK630 pKa = 9.98DD631 pKa = 3.31RR632 pKa = 11.84AGVSLIQRR640 pKa = 11.84PGLVQGAYY648 pKa = 8.55TPRR651 pKa = 11.84TLAAVCADD659 pKa = 3.18ASEE662 pKa = 4.12EE663 pKa = 3.92AGAFVAILFGSEE675 pKa = 4.28VQASWQQDD683 pKa = 3.63GMPVRR688 pKa = 11.84ADD690 pKa = 3.37ALQSSIEE697 pKa = 4.18ANCPDD702 pKa = 3.67PEE704 pKa = 4.44ARR706 pKa = 11.84QNVQALIDD714 pKa = 3.99ALQTPVTQPDD724 pKa = 3.57DD725 pKa = 3.8TVRR728 pKa = 11.84DD729 pKa = 3.71ALLQNANALIAGDD742 pKa = 3.67VTLEE746 pKa = 3.81QARR749 pKa = 11.84TGVEE753 pKa = 3.96DD754 pKa = 4.41ALNLYY759 pKa = 9.86LAEE762 pKa = 4.19QEE764 pKa = 4.22
Molecular weight: 80.63 kDa
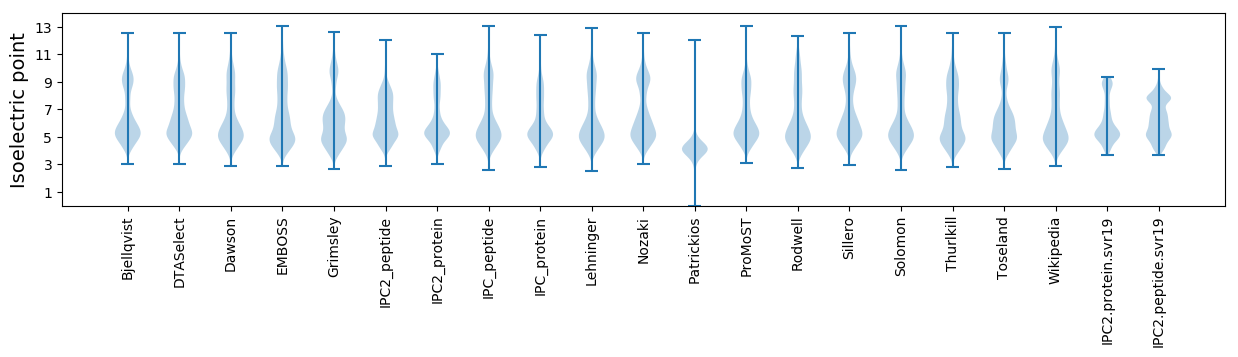
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4TER5|A0A1Y4TER5_9FIRM Ribonuclease P protein component OS=Gemmiger sp. An120 OX=1965549 GN=rnpA PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.27KK9 pKa = 7.52RR10 pKa = 11.84QRR12 pKa = 11.84SAVHH16 pKa = 6.23GFLNRR21 pKa = 11.84MSSRR25 pKa = 11.84SGRR28 pKa = 11.84KK29 pKa = 8.66VIKK32 pKa = 10.16ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84AKK41 pKa = 10.44LAAA44 pKa = 4.52
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.27KK9 pKa = 7.52RR10 pKa = 11.84QRR12 pKa = 11.84SAVHH16 pKa = 6.23GFLNRR21 pKa = 11.84MSSRR25 pKa = 11.84SGRR28 pKa = 11.84KK29 pKa = 8.66VIKK32 pKa = 10.16ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84AKK41 pKa = 10.44LAAA44 pKa = 4.52
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
991804 |
25 |
2252 |
325.4 |
35.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.614 ± 0.072 | 1.746 ± 0.018 |
5.403 ± 0.032 | 6.426 ± 0.038 |
3.806 ± 0.029 | 7.962 ± 0.042 |
1.719 ± 0.019 | 5.008 ± 0.041 |
4.034 ± 0.042 | 10.607 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.67 ± 0.022 | 3.092 ± 0.027 |
4.547 ± 0.03 | 3.827 ± 0.03 |
6.01 ± 0.045 | 5.137 ± 0.033 |
5.469 ± 0.031 | 7.191 ± 0.035 |
1.272 ± 0.019 | 3.459 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
