
Dongia mobilis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Dongia
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
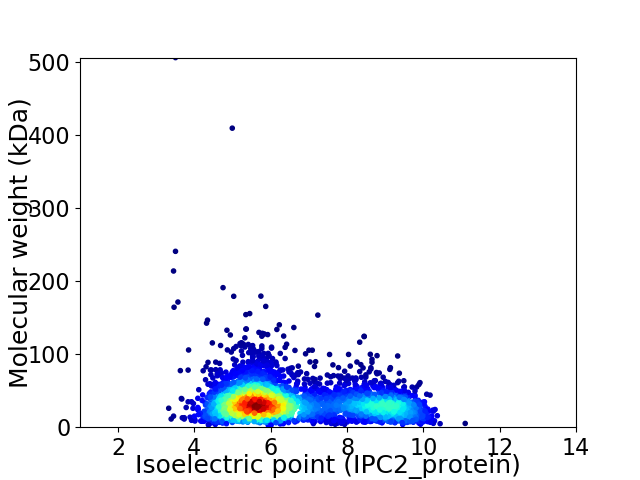
Virtual 2D-PAGE plot for 3804 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V3DF21|A0A4V3DF21_9PROT Tfp pilus assembly protein PilF OS=Dongia mobilis OX=578943 GN=A8950_0922 PE=4 SV=1
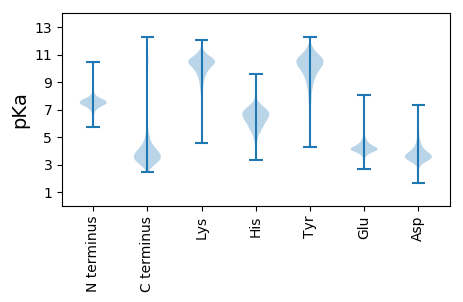
MM1 pKa = 7.12VPVAIANLGNLEE13 pKa = 4.15MKK15 pKa = 10.29KK16 pKa = 10.52VLLGTSALLGVGLLAGTAQASDD38 pKa = 3.89GVKK41 pKa = 10.51LSLGGYY47 pKa = 6.56MTNQMGVAFDD57 pKa = 4.69DD58 pKa = 4.76DD59 pKa = 4.39SAGEE63 pKa = 4.12AGDD66 pKa = 4.17NINAFGVGTDD76 pKa = 3.38AEE78 pKa = 4.69IYY80 pKa = 10.58FIGSVTLDD88 pKa = 3.1NGVTVGVRR96 pKa = 11.84FDD98 pKa = 4.9LEE100 pKa = 4.98GGDD103 pKa = 4.07EE104 pKa = 4.87NNDD107 pKa = 3.82QMDD110 pKa = 3.16QAYY113 pKa = 9.9AYY115 pKa = 9.73FKK117 pKa = 11.17GGFGDD122 pKa = 3.85IRR124 pKa = 11.84IGSQAGAAGNMYY136 pKa = 8.76MLPPGSTANFGPYY149 pKa = 9.84SPNTTGSALSPGVFDD164 pKa = 5.32PEE166 pKa = 4.42GMLANQDD173 pKa = 3.38KK174 pKa = 10.21SQKK177 pKa = 9.88LVYY180 pKa = 10.32YY181 pKa = 9.65SPNWSGFSFGVSFTPNDD198 pKa = 3.33NEE200 pKa = 4.0KK201 pKa = 10.99DD202 pKa = 3.88YY203 pKa = 11.79NNGDD207 pKa = 3.63NADD210 pKa = 4.02GQWRR214 pKa = 11.84AASAQGDD221 pKa = 3.76SDD223 pKa = 4.37NNIGLGLHH231 pKa = 5.58YY232 pKa = 9.55THH234 pKa = 7.17EE235 pKa = 4.85GDD237 pKa = 4.55GWGLDD242 pKa = 3.93LGAAAYY248 pKa = 9.24WEE250 pKa = 4.62GDD252 pKa = 3.6VQQKK256 pKa = 9.63VAGQDD261 pKa = 3.18EE262 pKa = 4.41QAGYY266 pKa = 9.87NAGINLSFGGFAFGVAGTYY285 pKa = 10.61LDD287 pKa = 4.89DD288 pKa = 5.1GNGAGQDD295 pKa = 2.75IWSIGTGMTYY305 pKa = 10.38NVEE308 pKa = 3.64AWTVGLGWAHH318 pKa = 7.25AEE320 pKa = 4.18LEE322 pKa = 4.48VAGARR327 pKa = 11.84DD328 pKa = 3.46ASADD332 pKa = 3.7RR333 pKa = 11.84VGVTGNYY340 pKa = 10.83AMGPGIDD347 pKa = 3.78LDD349 pKa = 4.12AGIFYY354 pKa = 9.0TWGEE358 pKa = 4.15DD359 pKa = 3.39AVGEE363 pKa = 4.04GDD365 pKa = 3.48YY366 pKa = 11.4DD367 pKa = 4.39SIEE370 pKa = 4.34FGVGSSISFF379 pKa = 3.54
MM1 pKa = 7.12VPVAIANLGNLEE13 pKa = 4.15MKK15 pKa = 10.29KK16 pKa = 10.52VLLGTSALLGVGLLAGTAQASDD38 pKa = 3.89GVKK41 pKa = 10.51LSLGGYY47 pKa = 6.56MTNQMGVAFDD57 pKa = 4.69DD58 pKa = 4.76DD59 pKa = 4.39SAGEE63 pKa = 4.12AGDD66 pKa = 4.17NINAFGVGTDD76 pKa = 3.38AEE78 pKa = 4.69IYY80 pKa = 10.58FIGSVTLDD88 pKa = 3.1NGVTVGVRR96 pKa = 11.84FDD98 pKa = 4.9LEE100 pKa = 4.98GGDD103 pKa = 4.07EE104 pKa = 4.87NNDD107 pKa = 3.82QMDD110 pKa = 3.16QAYY113 pKa = 9.9AYY115 pKa = 9.73FKK117 pKa = 11.17GGFGDD122 pKa = 3.85IRR124 pKa = 11.84IGSQAGAAGNMYY136 pKa = 8.76MLPPGSTANFGPYY149 pKa = 9.84SPNTTGSALSPGVFDD164 pKa = 5.32PEE166 pKa = 4.42GMLANQDD173 pKa = 3.38KK174 pKa = 10.21SQKK177 pKa = 9.88LVYY180 pKa = 10.32YY181 pKa = 9.65SPNWSGFSFGVSFTPNDD198 pKa = 3.33NEE200 pKa = 4.0KK201 pKa = 10.99DD202 pKa = 3.88YY203 pKa = 11.79NNGDD207 pKa = 3.63NADD210 pKa = 4.02GQWRR214 pKa = 11.84AASAQGDD221 pKa = 3.76SDD223 pKa = 4.37NNIGLGLHH231 pKa = 5.58YY232 pKa = 9.55THH234 pKa = 7.17EE235 pKa = 4.85GDD237 pKa = 4.55GWGLDD242 pKa = 3.93LGAAAYY248 pKa = 9.24WEE250 pKa = 4.62GDD252 pKa = 3.6VQQKK256 pKa = 9.63VAGQDD261 pKa = 3.18EE262 pKa = 4.41QAGYY266 pKa = 9.87NAGINLSFGGFAFGVAGTYY285 pKa = 10.61LDD287 pKa = 4.89DD288 pKa = 5.1GNGAGQDD295 pKa = 2.75IWSIGTGMTYY305 pKa = 10.38NVEE308 pKa = 3.64AWTVGLGWAHH318 pKa = 7.25AEE320 pKa = 4.18LEE322 pKa = 4.48VAGARR327 pKa = 11.84DD328 pKa = 3.46ASADD332 pKa = 3.7RR333 pKa = 11.84VGVTGNYY340 pKa = 10.83AMGPGIDD347 pKa = 3.78LDD349 pKa = 4.12AGIFYY354 pKa = 9.0TWGEE358 pKa = 4.15DD359 pKa = 3.39AVGEE363 pKa = 4.04GDD365 pKa = 3.48YY366 pKa = 11.4DD367 pKa = 4.39SIEE370 pKa = 4.34FGVGSSISFF379 pKa = 3.54
Molecular weight: 39.26 kDa
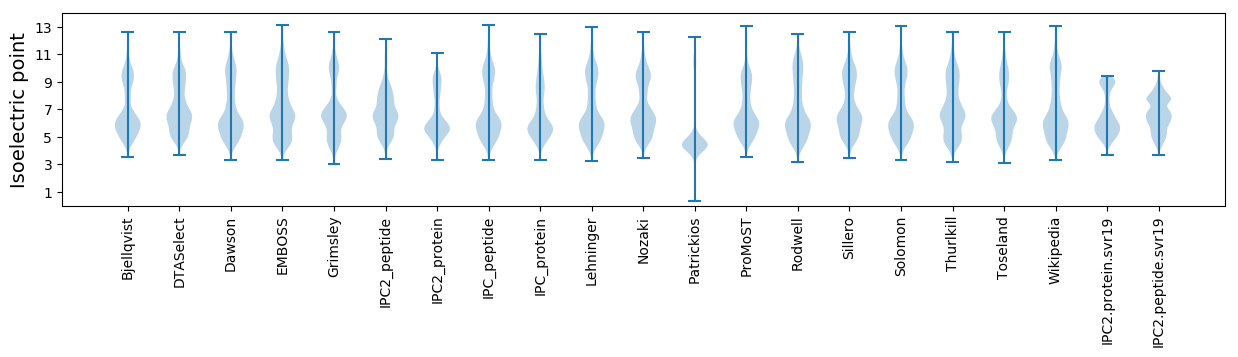
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6WN28|A0A4R6WN28_9PROT DNA helicase OS=Dongia mobilis OX=578943 GN=A8950_2245 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATNGGQKK29 pKa = 10.15VISSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.8GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATNGGQKK29 pKa = 10.15VISSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.8GRR39 pKa = 11.84KK40 pKa = 8.92RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1260373 |
27 |
4980 |
331.3 |
35.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.2 ± 0.064 | 0.886 ± 0.012 |
5.683 ± 0.042 | 5.538 ± 0.042 |
3.64 ± 0.024 | 8.824 ± 0.044 |
2.061 ± 0.019 | 5.361 ± 0.029 |
3.16 ± 0.032 | 10.651 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.478 ± 0.019 | 2.574 ± 0.027 |
5.219 ± 0.031 | 3.313 ± 0.026 |
7.144 ± 0.049 | 4.845 ± 0.026 |
4.807 ± 0.033 | 7.072 ± 0.032 |
1.343 ± 0.017 | 2.201 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
