
Circoviridae 16 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 8.64
Get precalculated fractions of proteins
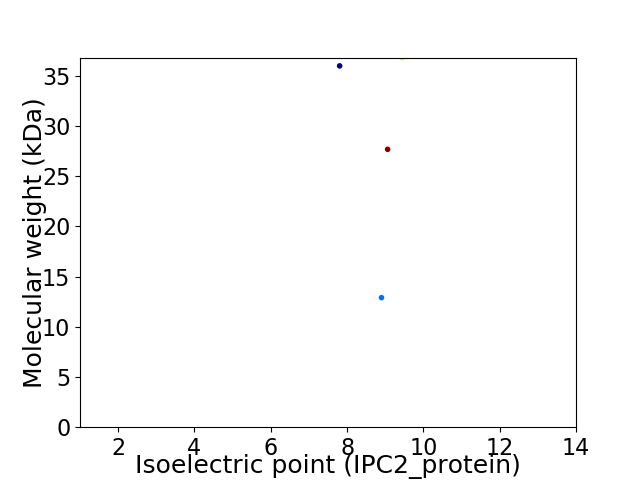
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
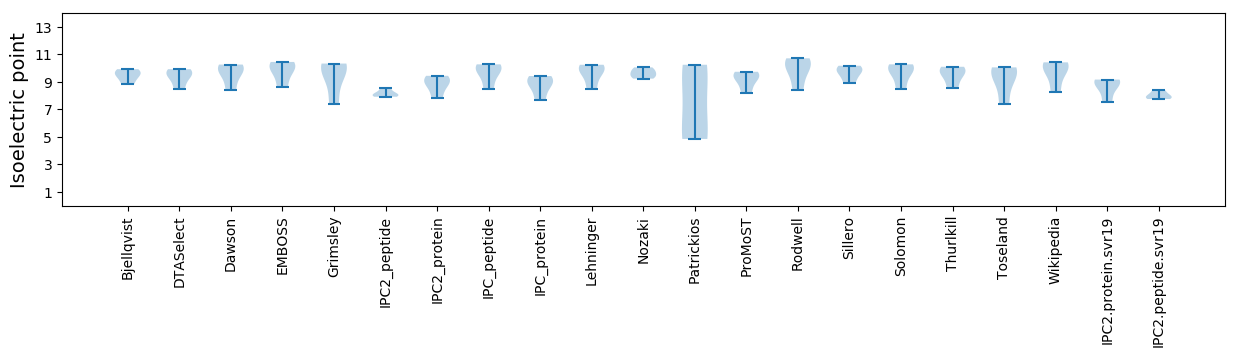
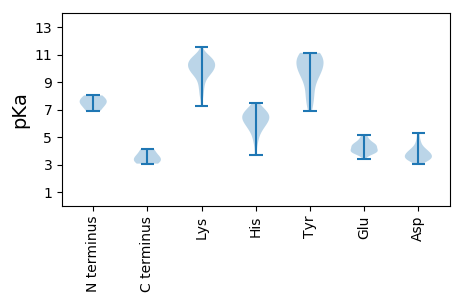
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5TNF0|S5TNF0_9CIRC Replication-associated protein OS=Circoviridae 16 LDMD-2013 OX=1379720 PE=4 SV=1
MM1 pKa = 7.54RR2 pKa = 11.84EE3 pKa = 3.75EE4 pKa = 3.89VRR6 pKa = 11.84IVGPLRR12 pKa = 11.84GPTLVSAVLITPAVNLHH29 pKa = 6.45DD30 pKa = 4.23VDD32 pKa = 3.29VHH34 pKa = 4.79TAHH37 pKa = 7.48RR38 pKa = 11.84GGVRR42 pKa = 11.84KK43 pKa = 9.58GHH45 pKa = 6.24DD46 pKa = 3.54TPDD49 pKa = 3.34LLRR52 pKa = 11.84IGDD55 pKa = 3.93VVVPIGVVACVVGGVAIRR73 pKa = 11.84TQVALKK79 pKa = 10.33RR80 pKa = 11.84HH81 pKa = 5.41VNLAGATNGLGARR94 pKa = 11.84VLVVQVEE101 pKa = 4.28VLGLKK106 pKa = 9.81DD107 pKa = 3.82VNVLTVEE114 pKa = 4.06VVPILRR120 pKa = 11.84IFEE123 pKa = 4.38SHH125 pKa = 5.79VSKK128 pKa = 10.0ATNRR132 pKa = 11.84FWKK135 pKa = 9.74AQNASTVYY143 pKa = 10.67EE144 pKa = 4.31SFLLANNVDD153 pKa = 5.15ANVEE157 pKa = 4.16WIRR160 pKa = 11.84TDD162 pKa = 4.17GEE164 pKa = 4.35VCMSLKK170 pKa = 10.18RR171 pKa = 11.84HH172 pKa = 5.5GRR174 pKa = 11.84RR175 pKa = 11.84QRR177 pKa = 11.84LVHH180 pKa = 6.4HH181 pKa = 6.69LAGGIVLNLKK191 pKa = 9.96VVVVLLVGGNVRR203 pKa = 11.84VVLLRR208 pKa = 11.84LCEE211 pKa = 3.85VRR213 pKa = 11.84NLVGTVVHH221 pKa = 6.77IGGPSGNTHH230 pKa = 7.0ASHH233 pKa = 6.87VLDD236 pKa = 3.86KK237 pKa = 11.54VVDD240 pKa = 4.1TEE242 pKa = 4.38THH244 pKa = 5.69GLLGIKK250 pKa = 9.85VRR252 pKa = 11.84TRR254 pKa = 11.84LKK256 pKa = 10.96AKK258 pKa = 9.98ILFAGLEE265 pKa = 4.01LAEE268 pKa = 4.53LCTLEE273 pKa = 4.92SLHH276 pKa = 7.02CSGEE280 pKa = 4.03LGDD283 pKa = 3.9ACGVLEE289 pKa = 4.19EE290 pKa = 4.98TGDD293 pKa = 3.18VWLALDD299 pKa = 4.79LRR301 pKa = 11.84DD302 pKa = 3.73GLKK305 pKa = 9.94TSTILCKK312 pKa = 10.03GTLSGSSLNCFNSTNTATSSXASSSLSS339 pKa = 3.35
MM1 pKa = 7.54RR2 pKa = 11.84EE3 pKa = 3.75EE4 pKa = 3.89VRR6 pKa = 11.84IVGPLRR12 pKa = 11.84GPTLVSAVLITPAVNLHH29 pKa = 6.45DD30 pKa = 4.23VDD32 pKa = 3.29VHH34 pKa = 4.79TAHH37 pKa = 7.48RR38 pKa = 11.84GGVRR42 pKa = 11.84KK43 pKa = 9.58GHH45 pKa = 6.24DD46 pKa = 3.54TPDD49 pKa = 3.34LLRR52 pKa = 11.84IGDD55 pKa = 3.93VVVPIGVVACVVGGVAIRR73 pKa = 11.84TQVALKK79 pKa = 10.33RR80 pKa = 11.84HH81 pKa = 5.41VNLAGATNGLGARR94 pKa = 11.84VLVVQVEE101 pKa = 4.28VLGLKK106 pKa = 9.81DD107 pKa = 3.82VNVLTVEE114 pKa = 4.06VVPILRR120 pKa = 11.84IFEE123 pKa = 4.38SHH125 pKa = 5.79VSKK128 pKa = 10.0ATNRR132 pKa = 11.84FWKK135 pKa = 9.74AQNASTVYY143 pKa = 10.67EE144 pKa = 4.31SFLLANNVDD153 pKa = 5.15ANVEE157 pKa = 4.16WIRR160 pKa = 11.84TDD162 pKa = 4.17GEE164 pKa = 4.35VCMSLKK170 pKa = 10.18RR171 pKa = 11.84HH172 pKa = 5.5GRR174 pKa = 11.84RR175 pKa = 11.84QRR177 pKa = 11.84LVHH180 pKa = 6.4HH181 pKa = 6.69LAGGIVLNLKK191 pKa = 9.96VVVVLLVGGNVRR203 pKa = 11.84VVLLRR208 pKa = 11.84LCEE211 pKa = 3.85VRR213 pKa = 11.84NLVGTVVHH221 pKa = 6.77IGGPSGNTHH230 pKa = 7.0ASHH233 pKa = 6.87VLDD236 pKa = 3.86KK237 pKa = 11.54VVDD240 pKa = 4.1TEE242 pKa = 4.38THH244 pKa = 5.69GLLGIKK250 pKa = 9.85VRR252 pKa = 11.84TRR254 pKa = 11.84LKK256 pKa = 10.96AKK258 pKa = 9.98ILFAGLEE265 pKa = 4.01LAEE268 pKa = 4.53LCTLEE273 pKa = 4.92SLHH276 pKa = 7.02CSGEE280 pKa = 4.03LGDD283 pKa = 3.9ACGVLEE289 pKa = 4.19EE290 pKa = 4.98TGDD293 pKa = 3.18VWLALDD299 pKa = 4.79LRR301 pKa = 11.84DD302 pKa = 3.73GLKK305 pKa = 9.94TSTILCKK312 pKa = 10.03GTLSGSSLNCFNSTNTATSSXASSSLSS339 pKa = 3.35
Molecular weight: 36.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5SYF3|S5SYF3_9CIRC Replication-associated protein OS=Circoviridae 16 LDMD-2013 OX=1379720 PE=4 SV=1
MM1 pKa = 8.03PYY3 pKa = 8.88GTSRR7 pKa = 11.84KK8 pKa = 8.12RR9 pKa = 11.84TARR12 pKa = 11.84XATSRR17 pKa = 11.84RR18 pKa = 11.84VSAVKK23 pKa = 9.56TIQAAARR30 pKa = 11.84KK31 pKa = 9.03RR32 pKa = 11.84AFTKK36 pKa = 10.57NRR38 pKa = 11.84GGLKK42 pKa = 10.0SVSKK46 pKa = 10.38IQRR49 pKa = 11.84QPYY52 pKa = 8.97VPRR55 pKa = 11.84LLKK58 pKa = 9.38NTASISQLSRR68 pKa = 11.84AVKK71 pKa = 9.62GLQRR75 pKa = 11.84AKK77 pKa = 10.59LGEE80 pKa = 4.0FQTGKK85 pKa = 10.35EE86 pKa = 3.97YY87 pKa = 11.11LGFQPSTYY95 pKa = 10.44FNAQKK100 pKa = 10.53PMCFCVNDD108 pKa = 3.96FVQYY112 pKa = 8.19VTGVGVPTWTTDD124 pKa = 3.26MNDD127 pKa = 3.6GANKK131 pKa = 9.88VSNFAKK137 pKa = 10.58AEE139 pKa = 4.11KK140 pKa = 10.36NDD142 pKa = 3.87TNVSSNEE149 pKa = 3.42KK150 pKa = 10.1NYY152 pKa = 10.72YY153 pKa = 8.62NFKK156 pKa = 10.34IQDD159 pKa = 3.49NTASKK164 pKa = 10.82VVYY167 pKa = 10.02QPLSSTMTFQAHH179 pKa = 6.46ANLSVGADD187 pKa = 3.62PLYY190 pKa = 10.92VRR192 pKa = 11.84IDD194 pKa = 3.06IVRR197 pKa = 11.84QKK199 pKa = 9.56KK200 pKa = 7.23TLVNGTRR207 pKa = 11.84VLRR210 pKa = 11.84LPEE213 pKa = 4.55SIGGLGNMALKK224 pKa = 10.49DD225 pKa = 3.48PKK227 pKa = 10.74YY228 pKa = 10.31RR229 pKa = 11.84NNFNSEE235 pKa = 4.4YY236 pKa = 10.43IHH238 pKa = 6.54VLQTKK243 pKa = 9.51YY244 pKa = 11.02LYY246 pKa = 11.08LNNKK250 pKa = 8.91DD251 pKa = 3.88AGAKK255 pKa = 8.71SVRR258 pKa = 11.84SSCKK262 pKa = 9.38IHH264 pKa = 6.96VPLQGYY270 pKa = 7.97LRR272 pKa = 11.84PDD274 pKa = 3.22GDD276 pKa = 3.82ATDD279 pKa = 3.62NAGNYY284 pKa = 7.58TDD286 pKa = 3.82WYY288 pKa = 11.12NNISDD293 pKa = 4.7SKK295 pKa = 10.86KK296 pKa = 8.45IWCIMSFSDD305 pKa = 3.61TSAMSGVDD313 pKa = 3.35INIMKK318 pKa = 9.35VNRR321 pKa = 11.84WRR323 pKa = 11.84DD324 pKa = 3.38QHH326 pKa = 6.12GTDD329 pKa = 3.55
MM1 pKa = 8.03PYY3 pKa = 8.88GTSRR7 pKa = 11.84KK8 pKa = 8.12RR9 pKa = 11.84TARR12 pKa = 11.84XATSRR17 pKa = 11.84RR18 pKa = 11.84VSAVKK23 pKa = 9.56TIQAAARR30 pKa = 11.84KK31 pKa = 9.03RR32 pKa = 11.84AFTKK36 pKa = 10.57NRR38 pKa = 11.84GGLKK42 pKa = 10.0SVSKK46 pKa = 10.38IQRR49 pKa = 11.84QPYY52 pKa = 8.97VPRR55 pKa = 11.84LLKK58 pKa = 9.38NTASISQLSRR68 pKa = 11.84AVKK71 pKa = 9.62GLQRR75 pKa = 11.84AKK77 pKa = 10.59LGEE80 pKa = 4.0FQTGKK85 pKa = 10.35EE86 pKa = 3.97YY87 pKa = 11.11LGFQPSTYY95 pKa = 10.44FNAQKK100 pKa = 10.53PMCFCVNDD108 pKa = 3.96FVQYY112 pKa = 8.19VTGVGVPTWTTDD124 pKa = 3.26MNDD127 pKa = 3.6GANKK131 pKa = 9.88VSNFAKK137 pKa = 10.58AEE139 pKa = 4.11KK140 pKa = 10.36NDD142 pKa = 3.87TNVSSNEE149 pKa = 3.42KK150 pKa = 10.1NYY152 pKa = 10.72YY153 pKa = 8.62NFKK156 pKa = 10.34IQDD159 pKa = 3.49NTASKK164 pKa = 10.82VVYY167 pKa = 10.02QPLSSTMTFQAHH179 pKa = 6.46ANLSVGADD187 pKa = 3.62PLYY190 pKa = 10.92VRR192 pKa = 11.84IDD194 pKa = 3.06IVRR197 pKa = 11.84QKK199 pKa = 9.56KK200 pKa = 7.23TLVNGTRR207 pKa = 11.84VLRR210 pKa = 11.84LPEE213 pKa = 4.55SIGGLGNMALKK224 pKa = 10.49DD225 pKa = 3.48PKK227 pKa = 10.74YY228 pKa = 10.31RR229 pKa = 11.84NNFNSEE235 pKa = 4.4YY236 pKa = 10.43IHH238 pKa = 6.54VLQTKK243 pKa = 9.51YY244 pKa = 11.02LYY246 pKa = 11.08LNNKK250 pKa = 8.91DD251 pKa = 3.88AGAKK255 pKa = 8.71SVRR258 pKa = 11.84SSCKK262 pKa = 9.38IHH264 pKa = 6.96VPLQGYY270 pKa = 7.97LRR272 pKa = 11.84PDD274 pKa = 3.22GDD276 pKa = 3.82ATDD279 pKa = 3.62NAGNYY284 pKa = 7.58TDD286 pKa = 3.82WYY288 pKa = 11.12NNISDD293 pKa = 4.7SKK295 pKa = 10.86KK296 pKa = 8.45IWCIMSFSDD305 pKa = 3.61TSAMSGVDD313 pKa = 3.35INIMKK318 pKa = 9.35VNRR321 pKa = 11.84WRR323 pKa = 11.84DD324 pKa = 3.38QHH326 pKa = 6.12GTDD329 pKa = 3.55
Molecular weight: 36.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1023 |
115 |
339 |
255.8 |
28.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.354 ± 0.498 | 1.271 ± 0.478 |
4.594 ± 0.528 | 4.399 ± 1.002 |
3.617 ± 0.992 | 7.82 ± 0.875 |
2.639 ± 0.657 | 3.812 ± 0.219 |
6.549 ± 1.102 | 8.7 ± 2.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.857 ± 0.581 | 5.572 ± 1.025 |
4.497 ± 1.253 | 3.519 ± 0.919 |
6.745 ± 0.448 | 7.136 ± 0.478 |
6.745 ± 0.27 | 9.384 ± 2.447 |
1.369 ± 0.257 | 3.03 ± 1.113 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
