
Boudabousia liubingyangii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Boudabousia
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
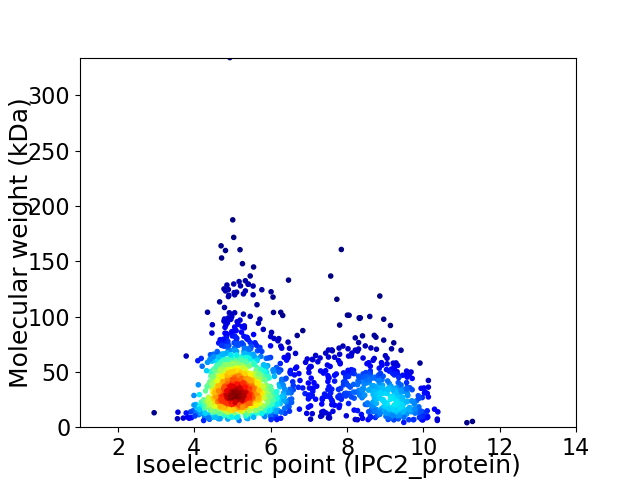
Virtual 2D-PAGE plot for 1617 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q5PQW1|A0A1Q5PQW1_9ACTO Uncharacterized protein OS=Boudabousia liubingyangii OX=1921764 GN=BSR29_02100 PE=4 SV=1
MM1 pKa = 7.72TDD3 pKa = 4.15FVPPFDD9 pKa = 4.82PALQALQNPNLDD21 pKa = 3.85PFVLAQIASDD31 pKa = 5.57RR32 pKa = 11.84PDD34 pKa = 3.08LWSTIVFHH42 pKa = 6.55PAAYY46 pKa = 9.24PEE48 pKa = 4.95LISYY52 pKa = 9.64LEE54 pKa = 4.02QYY56 pKa = 11.27GDD58 pKa = 3.66DD59 pKa = 3.79QVRR62 pKa = 11.84MAVQNRR68 pKa = 11.84RR69 pKa = 11.84NAEE72 pKa = 3.85AQGYY76 pKa = 7.05PLNYY80 pKa = 9.32PYY82 pKa = 10.77AAQPTMPPAPPAPPAAPAVPQYY104 pKa = 8.61PQPAQTFGTAPVRR117 pKa = 11.84RR118 pKa = 11.84QVFGTFEE125 pKa = 4.83HH126 pKa = 6.18NQQTYY131 pKa = 8.02PQPANEE137 pKa = 4.51PAPASYY143 pKa = 9.87EE144 pKa = 3.94AQPVSGEE151 pKa = 3.98PSPVGDD157 pKa = 3.56QVAPAGNEE165 pKa = 3.86FEE167 pKa = 5.62LPANDD172 pKa = 3.69ASQPFNEE179 pKa = 4.66AEE181 pKa = 4.12QPFNEE186 pKa = 4.47AEE188 pKa = 4.19QPVDD192 pKa = 4.36DD193 pKa = 4.36YY194 pKa = 12.11SEE196 pKa = 5.13LEE198 pKa = 3.92ATRR201 pKa = 11.84LVAHH205 pKa = 6.76PRR207 pKa = 11.84YY208 pKa = 10.19EE209 pKa = 4.15NVQALDD215 pKa = 3.68NEE217 pKa = 4.81TEE219 pKa = 3.86ATRR222 pKa = 11.84YY223 pKa = 8.76VPRR226 pKa = 11.84VTLPEE231 pKa = 4.65AEE233 pKa = 4.59GAQAPLPEE241 pKa = 5.74DD242 pKa = 3.65DD243 pKa = 3.96ATRR246 pKa = 11.84LVPRR250 pKa = 11.84VVLPAAEE257 pKa = 4.63DD258 pKa = 3.87GVYY261 pKa = 10.28VPSSVEE267 pKa = 3.64EE268 pKa = 4.4SPAPEE273 pKa = 4.86AEE275 pKa = 4.03AVPEE279 pKa = 4.22PEE281 pKa = 5.06AEE283 pKa = 4.22VAPEE287 pKa = 4.63LEE289 pKa = 4.68PEE291 pKa = 4.23AAPVPEE297 pKa = 5.01PEE299 pKa = 4.57SEE301 pKa = 4.31VAPEE305 pKa = 4.49PEE307 pKa = 4.52PEE309 pKa = 4.04AAPEE313 pKa = 3.84PVLEE317 pKa = 4.48PEE319 pKa = 4.66EE320 pKa = 4.2PASPEE325 pKa = 3.71VDD327 pKa = 3.34VVPEE331 pKa = 4.24PEE333 pKa = 4.37SEE335 pKa = 4.3VAPEE339 pKa = 4.26PEE341 pKa = 4.74PEE343 pKa = 4.35PEE345 pKa = 4.06VAPAPEE351 pKa = 4.59PEE353 pKa = 4.57PEE355 pKa = 4.32SEE357 pKa = 4.58AEE359 pKa = 3.79AAPAEE364 pKa = 4.41GEE366 pKa = 4.27FFVDD370 pKa = 5.37SIPSEE375 pKa = 4.62LNPQDD380 pKa = 4.83GEE382 pKa = 4.44TEE384 pKa = 4.11PDD386 pKa = 3.07HH387 pKa = 6.99GEE389 pKa = 3.9QSSEE393 pKa = 3.93EE394 pKa = 4.24SEE396 pKa = 4.71LEE398 pKa = 3.66SDD400 pKa = 4.15YY401 pKa = 11.88EE402 pKa = 3.99EE403 pKa = 6.19LEE405 pKa = 4.05ATRR408 pKa = 11.84VVPRR412 pKa = 11.84VHH414 pKa = 6.72KK415 pKa = 10.24PFIAPEE421 pKa = 4.04LQQSQGAPAPTPLEE435 pKa = 3.97VDD437 pKa = 3.8AAPAPSPGEE446 pKa = 4.04FEE448 pKa = 4.61AAPAPSPVDD457 pKa = 4.0FEE459 pKa = 4.46PAPAPSPEE467 pKa = 3.9AHH469 pKa = 6.13EE470 pKa = 4.5VAPAPGPEE478 pKa = 4.12DD479 pKa = 5.22LEE481 pKa = 4.49VAPAPSPADD490 pKa = 3.77YY491 pKa = 10.71EE492 pKa = 4.32PAPAPAPTEE501 pKa = 4.1YY502 pKa = 11.05APAPAPNPQYY512 pKa = 10.58EE513 pKa = 4.35PAPAPSGVMPPAFPDD528 pKa = 3.67VPQYY532 pKa = 10.93RR533 pKa = 11.84PNNEE537 pKa = 3.57EE538 pKa = 3.76TSQKK542 pKa = 10.58GSSKK546 pKa = 10.64RR547 pKa = 11.84KK548 pKa = 9.22LLWIILSVIVVIGLCVGGYY567 pKa = 9.45FLYY570 pKa = 10.46SYY572 pKa = 10.19LASGSDD578 pKa = 3.56NEE580 pKa = 4.51SLGNQATSQLQSEE593 pKa = 5.19YY594 pKa = 10.99VPTMYY599 pKa = 10.49FAA601 pKa = 5.55
MM1 pKa = 7.72TDD3 pKa = 4.15FVPPFDD9 pKa = 4.82PALQALQNPNLDD21 pKa = 3.85PFVLAQIASDD31 pKa = 5.57RR32 pKa = 11.84PDD34 pKa = 3.08LWSTIVFHH42 pKa = 6.55PAAYY46 pKa = 9.24PEE48 pKa = 4.95LISYY52 pKa = 9.64LEE54 pKa = 4.02QYY56 pKa = 11.27GDD58 pKa = 3.66DD59 pKa = 3.79QVRR62 pKa = 11.84MAVQNRR68 pKa = 11.84RR69 pKa = 11.84NAEE72 pKa = 3.85AQGYY76 pKa = 7.05PLNYY80 pKa = 9.32PYY82 pKa = 10.77AAQPTMPPAPPAPPAAPAVPQYY104 pKa = 8.61PQPAQTFGTAPVRR117 pKa = 11.84RR118 pKa = 11.84QVFGTFEE125 pKa = 4.83HH126 pKa = 6.18NQQTYY131 pKa = 8.02PQPANEE137 pKa = 4.51PAPASYY143 pKa = 9.87EE144 pKa = 3.94AQPVSGEE151 pKa = 3.98PSPVGDD157 pKa = 3.56QVAPAGNEE165 pKa = 3.86FEE167 pKa = 5.62LPANDD172 pKa = 3.69ASQPFNEE179 pKa = 4.66AEE181 pKa = 4.12QPFNEE186 pKa = 4.47AEE188 pKa = 4.19QPVDD192 pKa = 4.36DD193 pKa = 4.36YY194 pKa = 12.11SEE196 pKa = 5.13LEE198 pKa = 3.92ATRR201 pKa = 11.84LVAHH205 pKa = 6.76PRR207 pKa = 11.84YY208 pKa = 10.19EE209 pKa = 4.15NVQALDD215 pKa = 3.68NEE217 pKa = 4.81TEE219 pKa = 3.86ATRR222 pKa = 11.84YY223 pKa = 8.76VPRR226 pKa = 11.84VTLPEE231 pKa = 4.65AEE233 pKa = 4.59GAQAPLPEE241 pKa = 5.74DD242 pKa = 3.65DD243 pKa = 3.96ATRR246 pKa = 11.84LVPRR250 pKa = 11.84VVLPAAEE257 pKa = 4.63DD258 pKa = 3.87GVYY261 pKa = 10.28VPSSVEE267 pKa = 3.64EE268 pKa = 4.4SPAPEE273 pKa = 4.86AEE275 pKa = 4.03AVPEE279 pKa = 4.22PEE281 pKa = 5.06AEE283 pKa = 4.22VAPEE287 pKa = 4.63LEE289 pKa = 4.68PEE291 pKa = 4.23AAPVPEE297 pKa = 5.01PEE299 pKa = 4.57SEE301 pKa = 4.31VAPEE305 pKa = 4.49PEE307 pKa = 4.52PEE309 pKa = 4.04AAPEE313 pKa = 3.84PVLEE317 pKa = 4.48PEE319 pKa = 4.66EE320 pKa = 4.2PASPEE325 pKa = 3.71VDD327 pKa = 3.34VVPEE331 pKa = 4.24PEE333 pKa = 4.37SEE335 pKa = 4.3VAPEE339 pKa = 4.26PEE341 pKa = 4.74PEE343 pKa = 4.35PEE345 pKa = 4.06VAPAPEE351 pKa = 4.59PEE353 pKa = 4.57PEE355 pKa = 4.32SEE357 pKa = 4.58AEE359 pKa = 3.79AAPAEE364 pKa = 4.41GEE366 pKa = 4.27FFVDD370 pKa = 5.37SIPSEE375 pKa = 4.62LNPQDD380 pKa = 4.83GEE382 pKa = 4.44TEE384 pKa = 4.11PDD386 pKa = 3.07HH387 pKa = 6.99GEE389 pKa = 3.9QSSEE393 pKa = 3.93EE394 pKa = 4.24SEE396 pKa = 4.71LEE398 pKa = 3.66SDD400 pKa = 4.15YY401 pKa = 11.88EE402 pKa = 3.99EE403 pKa = 6.19LEE405 pKa = 4.05ATRR408 pKa = 11.84VVPRR412 pKa = 11.84VHH414 pKa = 6.72KK415 pKa = 10.24PFIAPEE421 pKa = 4.04LQQSQGAPAPTPLEE435 pKa = 3.97VDD437 pKa = 3.8AAPAPSPGEE446 pKa = 4.04FEE448 pKa = 4.61AAPAPSPVDD457 pKa = 4.0FEE459 pKa = 4.46PAPAPSPEE467 pKa = 3.9AHH469 pKa = 6.13EE470 pKa = 4.5VAPAPGPEE478 pKa = 4.12DD479 pKa = 5.22LEE481 pKa = 4.49VAPAPSPADD490 pKa = 3.77YY491 pKa = 10.71EE492 pKa = 4.32PAPAPAPTEE501 pKa = 4.1YY502 pKa = 11.05APAPAPNPQYY512 pKa = 10.58EE513 pKa = 4.35PAPAPSGVMPPAFPDD528 pKa = 3.67VPQYY532 pKa = 10.93RR533 pKa = 11.84PNNEE537 pKa = 3.57EE538 pKa = 3.76TSQKK542 pKa = 10.58GSSKK546 pKa = 10.64RR547 pKa = 11.84KK548 pKa = 9.22LLWIILSVIVVIGLCVGGYY567 pKa = 9.45FLYY570 pKa = 10.46SYY572 pKa = 10.19LASGSDD578 pKa = 3.56NEE580 pKa = 4.51SLGNQATSQLQSEE593 pKa = 5.19YY594 pKa = 10.99VPTMYY599 pKa = 10.49FAA601 pKa = 5.55
Molecular weight: 64.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q5PPC0|A0A1Q5PPC0_9ACTO Uncharacterized protein OS=Boudabousia liubingyangii OX=1921764 GN=BSR29_00315 PE=4 SV=1
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.4VHH17 pKa = 5.52GFRR20 pKa = 11.84KK21 pKa = 9.95RR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.67GRR40 pKa = 11.84ASISAA45 pKa = 3.59
MM1 pKa = 7.41VKK3 pKa = 9.03RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.4VHH17 pKa = 5.52GFRR20 pKa = 11.84KK21 pKa = 9.95RR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.67GRR40 pKa = 11.84ASISAA45 pKa = 3.59
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
587279 |
32 |
3168 |
363.2 |
39.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.064 ± 0.087 | 0.745 ± 0.019 |
5.289 ± 0.05 | 6.494 ± 0.059 |
3.508 ± 0.038 | 7.876 ± 0.056 |
1.882 ± 0.023 | 5.068 ± 0.055 |
4.335 ± 0.055 | 10.35 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.094 ± 0.026 | 3.201 ± 0.038 |
5.384 ± 0.06 | 4.22 ± 0.045 |
5.46 ± 0.058 | 5.835 ± 0.042 |
5.725 ± 0.048 | 7.505 ± 0.059 |
1.446 ± 0.026 | 2.519 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
