
Fringilla coelebs papillomavirus (isolate Chaffinch/Netherlands/Dutch) (FcPV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Etapapillomavirus; Etapapillomavirus 1
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
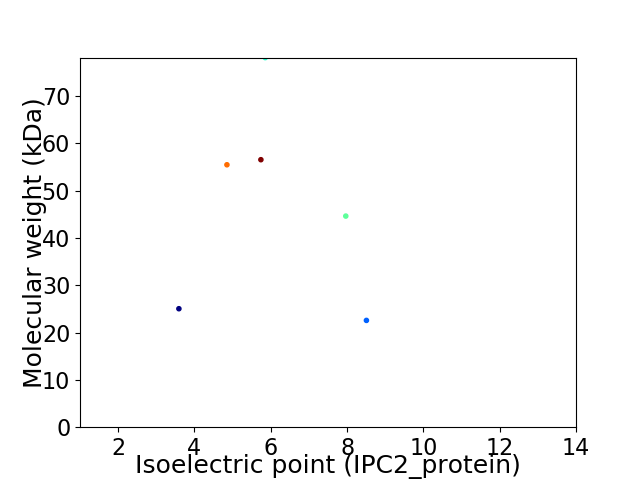
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8JNA7|Q8JNA7_FCPVN Putative E7 protein OS=Fringilla coelebs papillomavirus (isolate Chaffinch/Netherlands/Dutch) OX=654914 GN=E7 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84NRR4 pKa = 11.84LPIGAQGPPGGQPSNNPSEE23 pKa = 5.26NNFNPEE29 pKa = 3.8DD30 pKa = 3.88WDD32 pKa = 3.67ILLDD36 pKa = 3.8TDD38 pKa = 4.1SSSGSEE44 pKa = 4.21TEE46 pKa = 4.09TAEE49 pKa = 3.98ATSPSSSEE57 pKa = 3.72EE58 pKa = 3.77SAEE61 pKa = 3.96FDD63 pKa = 3.55WEE65 pKa = 4.22VKK67 pKa = 10.78LIVTGNSQDD76 pKa = 4.34PITLQEE82 pKa = 4.77LEE84 pKa = 5.1DD85 pKa = 4.12LNQVLVAEE93 pKa = 5.25LGHH96 pKa = 6.87PATVYY101 pKa = 10.56SIQDD105 pKa = 3.39QADD108 pKa = 3.88GASYY112 pKa = 10.38PPGSPTPSTSTFPFEE127 pKa = 4.29TPQGNNQFTSMAVAGSSAAADD148 pKa = 3.75PPSFPSPASVVDD160 pKa = 4.68LVCHH164 pKa = 6.35EE165 pKa = 4.65SMGDD169 pKa = 3.25SDD171 pKa = 4.1VDD173 pKa = 3.99EE174 pKa = 5.24EE175 pKa = 4.34EE176 pKa = 4.76HH177 pKa = 7.2LPNNPANTPEE187 pKa = 3.86EE188 pKa = 4.27SGANDD193 pKa = 3.59TEE195 pKa = 4.89FKK197 pKa = 10.35CTICSKK203 pKa = 10.35PLTEE207 pKa = 5.31GEE209 pKa = 3.91LDD211 pKa = 3.07EE212 pKa = 4.79WGLVQGDD219 pKa = 3.5EE220 pKa = 4.58GLCHH224 pKa = 6.49FCGFGAGVVDD234 pKa = 5.76FFPP237 pKa = 5.61
MM1 pKa = 8.06RR2 pKa = 11.84NRR4 pKa = 11.84LPIGAQGPPGGQPSNNPSEE23 pKa = 5.26NNFNPEE29 pKa = 3.8DD30 pKa = 3.88WDD32 pKa = 3.67ILLDD36 pKa = 3.8TDD38 pKa = 4.1SSSGSEE44 pKa = 4.21TEE46 pKa = 4.09TAEE49 pKa = 3.98ATSPSSSEE57 pKa = 3.72EE58 pKa = 3.77SAEE61 pKa = 3.96FDD63 pKa = 3.55WEE65 pKa = 4.22VKK67 pKa = 10.78LIVTGNSQDD76 pKa = 4.34PITLQEE82 pKa = 4.77LEE84 pKa = 5.1DD85 pKa = 4.12LNQVLVAEE93 pKa = 5.25LGHH96 pKa = 6.87PATVYY101 pKa = 10.56SIQDD105 pKa = 3.39QADD108 pKa = 3.88GASYY112 pKa = 10.38PPGSPTPSTSTFPFEE127 pKa = 4.29TPQGNNQFTSMAVAGSSAAADD148 pKa = 3.75PPSFPSPASVVDD160 pKa = 4.68LVCHH164 pKa = 6.35EE165 pKa = 4.65SMGDD169 pKa = 3.25SDD171 pKa = 4.1VDD173 pKa = 3.99EE174 pKa = 5.24EE175 pKa = 4.34EE176 pKa = 4.76HH177 pKa = 7.2LPNNPANTPEE187 pKa = 3.86EE188 pKa = 4.27SGANDD193 pKa = 3.59TEE195 pKa = 4.89FKK197 pKa = 10.35CTICSKK203 pKa = 10.35PLTEE207 pKa = 5.31GEE209 pKa = 3.91LDD211 pKa = 3.07EE212 pKa = 4.79WGLVQGDD219 pKa = 3.5EE220 pKa = 4.58GLCHH224 pKa = 6.49FCGFGAGVVDD234 pKa = 5.76FFPP237 pKa = 5.61
Molecular weight: 25.04 kDa
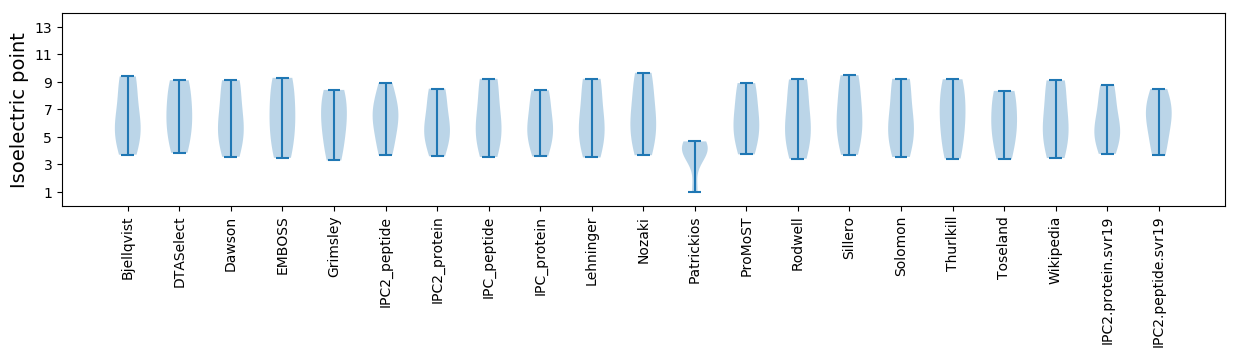
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8JNA6|Q8JNA6_FCPVN Putative replication protein E1 OS=Fringilla coelebs papillomavirus (isolate Chaffinch/Netherlands/Dutch) OX=654914 GN=E1 PE=4 SV=1
MM1 pKa = 7.48EE2 pKa = 5.89LSNPQGLPQTDD13 pKa = 3.46RR14 pKa = 11.84CIGSCCSSNSSSSGMRR30 pKa = 11.84LTLFCGSWCKK40 pKa = 10.52VVAIGNGGHH49 pKa = 7.5LPRR52 pKa = 11.84IPMTQLLISTISRR65 pKa = 11.84AQCRR69 pKa = 11.84KK70 pKa = 9.5LCTAAPYY77 pKa = 10.71LFLKK81 pKa = 10.43QCVCIHH87 pKa = 6.77RR88 pKa = 11.84MHH90 pKa = 7.02CLLPIFSQLKK100 pKa = 9.3RR101 pKa = 11.84ICAGLPPEE109 pKa = 4.46GWLGTVADD117 pKa = 4.63KK118 pKa = 10.5LTMFICQMQNEE129 pKa = 4.64CLKK132 pKa = 10.18HH133 pKa = 5.74YY134 pKa = 9.92PYY136 pKa = 9.15EE137 pKa = 3.98TLEE140 pKa = 3.64IVMPYY145 pKa = 8.58TALRR149 pKa = 11.84EE150 pKa = 4.39LYY152 pKa = 10.65SHH154 pKa = 6.94LQLTASQRR162 pKa = 11.84RR163 pKa = 11.84TKK165 pKa = 10.73VSLILRR171 pKa = 11.84DD172 pKa = 3.32IRR174 pKa = 11.84HH175 pKa = 5.7CQLVRR180 pKa = 11.84EE181 pKa = 4.82SKK183 pKa = 9.77HH184 pKa = 3.94QARR187 pKa = 11.84GRR189 pKa = 11.84KK190 pKa = 7.61DD191 pKa = 2.89HH192 pKa = 7.08RR193 pKa = 11.84NYY195 pKa = 10.52FNAAA199 pKa = 3.23
MM1 pKa = 7.48EE2 pKa = 5.89LSNPQGLPQTDD13 pKa = 3.46RR14 pKa = 11.84CIGSCCSSNSSSSGMRR30 pKa = 11.84LTLFCGSWCKK40 pKa = 10.52VVAIGNGGHH49 pKa = 7.5LPRR52 pKa = 11.84IPMTQLLISTISRR65 pKa = 11.84AQCRR69 pKa = 11.84KK70 pKa = 9.5LCTAAPYY77 pKa = 10.71LFLKK81 pKa = 10.43QCVCIHH87 pKa = 6.77RR88 pKa = 11.84MHH90 pKa = 7.02CLLPIFSQLKK100 pKa = 9.3RR101 pKa = 11.84ICAGLPPEE109 pKa = 4.46GWLGTVADD117 pKa = 4.63KK118 pKa = 10.5LTMFICQMQNEE129 pKa = 4.64CLKK132 pKa = 10.18HH133 pKa = 5.74YY134 pKa = 9.92PYY136 pKa = 9.15EE137 pKa = 3.98TLEE140 pKa = 3.64IVMPYY145 pKa = 8.58TALRR149 pKa = 11.84EE150 pKa = 4.39LYY152 pKa = 10.65SHH154 pKa = 6.94LQLTASQRR162 pKa = 11.84RR163 pKa = 11.84TKK165 pKa = 10.73VSLILRR171 pKa = 11.84DD172 pKa = 3.32IRR174 pKa = 11.84HH175 pKa = 5.7CQLVRR180 pKa = 11.84EE181 pKa = 4.82SKK183 pKa = 9.77HH184 pKa = 3.94QARR187 pKa = 11.84GRR189 pKa = 11.84KK190 pKa = 7.61DD191 pKa = 2.89HH192 pKa = 7.08RR193 pKa = 11.84NYY195 pKa = 10.52FNAAA199 pKa = 3.23
Molecular weight: 22.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2540 |
199 |
694 |
423.3 |
47.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.535 ± 0.366 | 2.047 ± 0.548 |
5.906 ± 0.338 | 6.339 ± 0.527 |
3.386 ± 0.36 | 7.205 ± 0.767 |
2.283 ± 0.387 | 4.291 ± 0.411 |
3.858 ± 0.87 | 8.583 ± 0.67 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.693 ± 0.176 | 4.409 ± 0.736 |
7.087 ± 0.936 | 4.173 ± 0.298 |
6.378 ± 0.767 | 8.543 ± 0.763 |
5.827 ± 0.221 | 6.535 ± 0.736 |
1.535 ± 0.122 | 3.386 ± 0.419 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
