
Daphnis nerii cypovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Cypovirus; unclassified Cypovirus
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
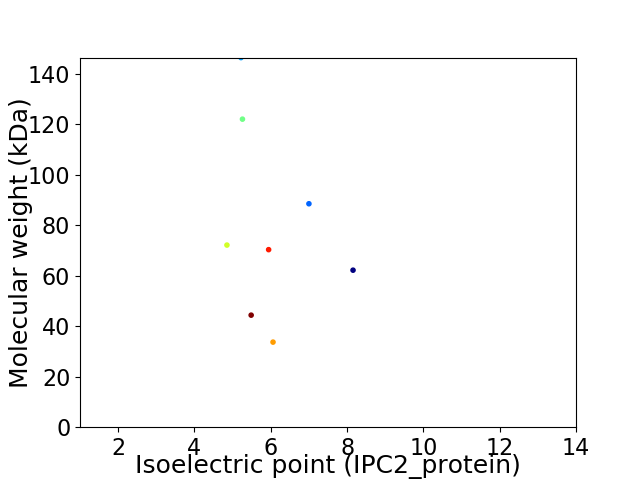
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385LMP6|A0A385LMP6_9REOV Uncharacterized protein OS=Daphnis nerii cypovirus OX=1986950 PE=4 SV=1
MM1 pKa = 7.27MLYY4 pKa = 10.47AIDD7 pKa = 4.19VLNKK11 pKa = 8.54ATKK14 pKa = 9.68TIQQLRR20 pKa = 11.84VFKK23 pKa = 11.06VDD25 pKa = 3.35GQSTDD30 pKa = 3.77DD31 pKa = 3.34IRR33 pKa = 11.84ALRR36 pKa = 11.84TEE38 pKa = 4.44FEE40 pKa = 4.21RR41 pKa = 11.84LTGKK45 pKa = 10.5KK46 pKa = 9.84LLSRR50 pKa = 11.84KK51 pKa = 7.54YY52 pKa = 9.16TSALTSKK59 pKa = 10.73LFDD62 pKa = 3.47EE63 pKa = 5.23SVTKK67 pKa = 10.89NSNNKK72 pKa = 8.08LTYY75 pKa = 9.99HH76 pKa = 6.47IQNKK80 pKa = 9.73SMIGTHH86 pKa = 6.58LLSLDD91 pKa = 3.46TDD93 pKa = 3.92KK94 pKa = 11.65VIWTSAEE101 pKa = 4.1DD102 pKa = 3.49IKK104 pKa = 11.66SKK106 pKa = 9.93ICEE109 pKa = 4.28SEE111 pKa = 4.1VTLQSIKK118 pKa = 10.33MIEE121 pKa = 4.07SDD123 pKa = 3.69LTKK126 pKa = 10.57ALQMYY131 pKa = 10.51DD132 pKa = 3.23DD133 pKa = 4.11TAPTKK138 pKa = 10.01TVISADD144 pKa = 3.47EE145 pKa = 4.64RR146 pKa = 11.84SVSWKK151 pKa = 9.14TDD153 pKa = 2.83PKK155 pKa = 9.5MIKK158 pKa = 10.06VVIRR162 pKa = 11.84DD163 pKa = 3.97GKK165 pKa = 10.69DD166 pKa = 3.24EE167 pKa = 4.53SCFGLGTEE175 pKa = 5.16KK176 pKa = 10.52IPMRR180 pKa = 11.84DD181 pKa = 3.08SNFYY185 pKa = 10.67ANEE188 pKa = 3.76YY189 pKa = 10.66DD190 pKa = 4.62LEE192 pKa = 4.44QNMMASVNFLNVASSFDD209 pKa = 3.65DD210 pKa = 3.86SKK212 pKa = 11.75SLKK215 pKa = 10.43LKK217 pKa = 10.67SDD219 pKa = 3.62LAKK222 pKa = 10.89VGTAYY227 pKa = 9.46EE228 pKa = 4.36QWCNDD233 pKa = 3.1HH234 pKa = 6.82PQISDD239 pKa = 3.3QYY241 pKa = 7.33QVRR244 pKa = 11.84MIVRR248 pKa = 11.84ASTYY252 pKa = 10.17EE253 pKa = 3.77EE254 pKa = 4.34PLIKK258 pKa = 10.58DD259 pKa = 3.87LLSNIASTCIFTCAIYY275 pKa = 10.98NEE277 pKa = 4.68DD278 pKa = 3.86GTWSVTDD285 pKa = 4.08SFNDD289 pKa = 3.33TDD291 pKa = 3.9TVLAPVEE298 pKa = 4.24EE299 pKa = 4.81LSATANWSTPEE310 pKa = 4.11TVDD313 pKa = 3.59GQAEE317 pKa = 4.23QQVVASEE324 pKa = 4.77SIASATATTEE334 pKa = 4.33SNVDD338 pKa = 3.28TAINEE343 pKa = 4.05QAVEE347 pKa = 4.05INDD350 pKa = 3.4VDD352 pKa = 3.72EE353 pKa = 4.7STTEE357 pKa = 3.75ASQDD361 pKa = 2.96ATVIEE366 pKa = 4.33KK367 pKa = 10.31PLVEE371 pKa = 4.07EE372 pKa = 4.24EE373 pKa = 4.05QRR375 pKa = 11.84SVDD378 pKa = 3.38TTHH381 pKa = 7.95RR382 pKa = 11.84IQSVEE387 pKa = 3.91KK388 pKa = 9.38TVNAVPTKK396 pKa = 10.0PCEE399 pKa = 3.9KK400 pKa = 10.65DD401 pKa = 3.12EE402 pKa = 4.68LSNEE406 pKa = 3.85KK407 pKa = 8.97KK408 pKa = 9.24TLIRR412 pKa = 11.84LIGSEE417 pKa = 4.33LDD419 pKa = 3.19GDD421 pKa = 3.92KK422 pKa = 11.04PIVVNVASDD431 pKa = 3.6YY432 pKa = 10.54TLQDD436 pKa = 3.4QIEE439 pKa = 4.1DD440 pKa = 4.45LINNKK445 pKa = 7.49TANIRR450 pKa = 11.84IWHH453 pKa = 5.62NEE455 pKa = 3.2ILIGRR460 pKa = 11.84RR461 pKa = 11.84DD462 pKa = 3.93TIHH465 pKa = 6.9SLRR468 pKa = 11.84AGNLSEE474 pKa = 5.94FIIKK478 pKa = 9.64FQTITIPEE486 pKa = 4.44SNVPTFNVKK495 pKa = 10.53LNTGSNIGVEE505 pKa = 4.0PFAVKK510 pKa = 10.81NNTTRR515 pKa = 11.84SSNDD519 pKa = 3.05EE520 pKa = 3.86LFEE523 pKa = 4.39WGFKK527 pKa = 10.22TMVEE531 pKa = 4.28SLIKK535 pKa = 8.8THH537 pKa = 7.36LSFLHH542 pKa = 5.77STNPTADD549 pKa = 4.21RR550 pKa = 11.84IIITYY555 pKa = 8.3GKK557 pKa = 9.94KK558 pKa = 10.34YY559 pKa = 10.33KK560 pKa = 10.62ADD562 pKa = 3.83DD563 pKa = 4.11FEE565 pKa = 6.89LEE567 pKa = 4.2MVNGFRR573 pKa = 11.84VMTHH577 pKa = 6.14KK578 pKa = 10.42KK579 pKa = 10.31SKK581 pKa = 9.96ISAFKK586 pKa = 11.05SEE588 pKa = 4.43TIEE591 pKa = 4.43NLLKK595 pKa = 10.32MAGDD599 pKa = 4.45DD600 pKa = 3.51EE601 pKa = 5.19AKK603 pKa = 10.67RR604 pKa = 11.84NMIYY608 pKa = 10.73DD609 pKa = 3.85VIDD612 pKa = 4.73NIMSSCLSEE621 pKa = 5.04AEE623 pKa = 4.1SPYY626 pKa = 10.71TSFQQRR632 pKa = 11.84HH633 pKa = 5.35LNVIGLMRR641 pKa = 11.84DD642 pKa = 3.03
MM1 pKa = 7.27MLYY4 pKa = 10.47AIDD7 pKa = 4.19VLNKK11 pKa = 8.54ATKK14 pKa = 9.68TIQQLRR20 pKa = 11.84VFKK23 pKa = 11.06VDD25 pKa = 3.35GQSTDD30 pKa = 3.77DD31 pKa = 3.34IRR33 pKa = 11.84ALRR36 pKa = 11.84TEE38 pKa = 4.44FEE40 pKa = 4.21RR41 pKa = 11.84LTGKK45 pKa = 10.5KK46 pKa = 9.84LLSRR50 pKa = 11.84KK51 pKa = 7.54YY52 pKa = 9.16TSALTSKK59 pKa = 10.73LFDD62 pKa = 3.47EE63 pKa = 5.23SVTKK67 pKa = 10.89NSNNKK72 pKa = 8.08LTYY75 pKa = 9.99HH76 pKa = 6.47IQNKK80 pKa = 9.73SMIGTHH86 pKa = 6.58LLSLDD91 pKa = 3.46TDD93 pKa = 3.92KK94 pKa = 11.65VIWTSAEE101 pKa = 4.1DD102 pKa = 3.49IKK104 pKa = 11.66SKK106 pKa = 9.93ICEE109 pKa = 4.28SEE111 pKa = 4.1VTLQSIKK118 pKa = 10.33MIEE121 pKa = 4.07SDD123 pKa = 3.69LTKK126 pKa = 10.57ALQMYY131 pKa = 10.51DD132 pKa = 3.23DD133 pKa = 4.11TAPTKK138 pKa = 10.01TVISADD144 pKa = 3.47EE145 pKa = 4.64RR146 pKa = 11.84SVSWKK151 pKa = 9.14TDD153 pKa = 2.83PKK155 pKa = 9.5MIKK158 pKa = 10.06VVIRR162 pKa = 11.84DD163 pKa = 3.97GKK165 pKa = 10.69DD166 pKa = 3.24EE167 pKa = 4.53SCFGLGTEE175 pKa = 5.16KK176 pKa = 10.52IPMRR180 pKa = 11.84DD181 pKa = 3.08SNFYY185 pKa = 10.67ANEE188 pKa = 3.76YY189 pKa = 10.66DD190 pKa = 4.62LEE192 pKa = 4.44QNMMASVNFLNVASSFDD209 pKa = 3.65DD210 pKa = 3.86SKK212 pKa = 11.75SLKK215 pKa = 10.43LKK217 pKa = 10.67SDD219 pKa = 3.62LAKK222 pKa = 10.89VGTAYY227 pKa = 9.46EE228 pKa = 4.36QWCNDD233 pKa = 3.1HH234 pKa = 6.82PQISDD239 pKa = 3.3QYY241 pKa = 7.33QVRR244 pKa = 11.84MIVRR248 pKa = 11.84ASTYY252 pKa = 10.17EE253 pKa = 3.77EE254 pKa = 4.34PLIKK258 pKa = 10.58DD259 pKa = 3.87LLSNIASTCIFTCAIYY275 pKa = 10.98NEE277 pKa = 4.68DD278 pKa = 3.86GTWSVTDD285 pKa = 4.08SFNDD289 pKa = 3.33TDD291 pKa = 3.9TVLAPVEE298 pKa = 4.24EE299 pKa = 4.81LSATANWSTPEE310 pKa = 4.11TVDD313 pKa = 3.59GQAEE317 pKa = 4.23QQVVASEE324 pKa = 4.77SIASATATTEE334 pKa = 4.33SNVDD338 pKa = 3.28TAINEE343 pKa = 4.05QAVEE347 pKa = 4.05INDD350 pKa = 3.4VDD352 pKa = 3.72EE353 pKa = 4.7STTEE357 pKa = 3.75ASQDD361 pKa = 2.96ATVIEE366 pKa = 4.33KK367 pKa = 10.31PLVEE371 pKa = 4.07EE372 pKa = 4.24EE373 pKa = 4.05QRR375 pKa = 11.84SVDD378 pKa = 3.38TTHH381 pKa = 7.95RR382 pKa = 11.84IQSVEE387 pKa = 3.91KK388 pKa = 9.38TVNAVPTKK396 pKa = 10.0PCEE399 pKa = 3.9KK400 pKa = 10.65DD401 pKa = 3.12EE402 pKa = 4.68LSNEE406 pKa = 3.85KK407 pKa = 8.97KK408 pKa = 9.24TLIRR412 pKa = 11.84LIGSEE417 pKa = 4.33LDD419 pKa = 3.19GDD421 pKa = 3.92KK422 pKa = 11.04PIVVNVASDD431 pKa = 3.6YY432 pKa = 10.54TLQDD436 pKa = 3.4QIEE439 pKa = 4.1DD440 pKa = 4.45LINNKK445 pKa = 7.49TANIRR450 pKa = 11.84IWHH453 pKa = 5.62NEE455 pKa = 3.2ILIGRR460 pKa = 11.84RR461 pKa = 11.84DD462 pKa = 3.93TIHH465 pKa = 6.9SLRR468 pKa = 11.84AGNLSEE474 pKa = 5.94FIIKK478 pKa = 9.64FQTITIPEE486 pKa = 4.44SNVPTFNVKK495 pKa = 10.53LNTGSNIGVEE505 pKa = 4.0PFAVKK510 pKa = 10.81NNTTRR515 pKa = 11.84SSNDD519 pKa = 3.05EE520 pKa = 3.86LFEE523 pKa = 4.39WGFKK527 pKa = 10.22TMVEE531 pKa = 4.28SLIKK535 pKa = 8.8THH537 pKa = 7.36LSFLHH542 pKa = 5.77STNPTADD549 pKa = 4.21RR550 pKa = 11.84IIITYY555 pKa = 8.3GKK557 pKa = 9.94KK558 pKa = 10.34YY559 pKa = 10.33KK560 pKa = 10.62ADD562 pKa = 3.83DD563 pKa = 4.11FEE565 pKa = 6.89LEE567 pKa = 4.2MVNGFRR573 pKa = 11.84VMTHH577 pKa = 6.14KK578 pKa = 10.42KK579 pKa = 10.31SKK581 pKa = 9.96ISAFKK586 pKa = 11.05SEE588 pKa = 4.43TIEE591 pKa = 4.43NLLKK595 pKa = 10.32MAGDD599 pKa = 4.45DD600 pKa = 3.51EE601 pKa = 5.19AKK603 pKa = 10.67RR604 pKa = 11.84NMIYY608 pKa = 10.73DD609 pKa = 3.85VIDD612 pKa = 4.73NIMSSCLSEE621 pKa = 5.04AEE623 pKa = 4.1SPYY626 pKa = 10.71TSFQQRR632 pKa = 11.84HH633 pKa = 5.35LNVIGLMRR641 pKa = 11.84DD642 pKa = 3.03
Molecular weight: 72.18 kDa
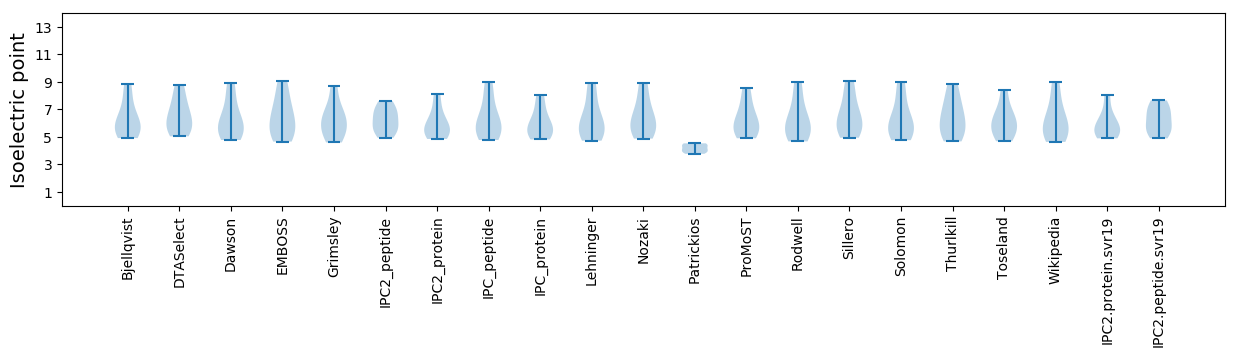
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385LMP0|A0A385LMP0_9REOV Uncharacterized protein OS=Daphnis nerii cypovirus OX=1986950 PE=4 SV=1
MM1 pKa = 7.7LFTINIRR8 pKa = 11.84APKK11 pKa = 9.14TLATLNRR18 pKa = 11.84MGIYY22 pKa = 10.24LRR24 pKa = 11.84EE25 pKa = 4.04QYY27 pKa = 10.64KK28 pKa = 10.42HH29 pKa = 6.13EE30 pKa = 4.28KK31 pKa = 8.72TSKK34 pKa = 10.77LLLKK38 pKa = 10.78RR39 pKa = 11.84NDD41 pKa = 4.13EE42 pKa = 4.02LHH44 pKa = 6.43HH45 pKa = 5.9WLEE48 pKa = 4.39KK49 pKa = 10.56LSARR53 pKa = 11.84HH54 pKa = 5.96KK55 pKa = 9.81GHH57 pKa = 6.14VFSMDD62 pKa = 2.66AVARR66 pKa = 11.84ILTEE70 pKa = 3.72RR71 pKa = 11.84LMKK74 pKa = 10.38INIIDD79 pKa = 3.57KK80 pKa = 10.71RR81 pKa = 11.84LGKK84 pKa = 10.74VKK86 pKa = 10.53DD87 pKa = 3.24IFANEE92 pKa = 4.38SKK94 pKa = 11.0NILQIVNKK102 pKa = 9.95EE103 pKa = 4.15DD104 pKa = 3.39VDD106 pKa = 4.47GVAMTYY112 pKa = 11.19DD113 pKa = 3.68DD114 pKa = 5.53ASFLLTLTKK123 pKa = 10.47EE124 pKa = 4.26VVIPNNEE131 pKa = 3.95TPVLYY136 pKa = 10.59EE137 pKa = 3.98IDD139 pKa = 4.19HH140 pKa = 6.12PTLRR144 pKa = 11.84HH145 pKa = 6.55LIPSYY150 pKa = 9.64IDD152 pKa = 3.04QMRR155 pKa = 11.84KK156 pKa = 8.42VLIIRR161 pKa = 11.84RR162 pKa = 11.84VTITDD167 pKa = 3.34VLYY170 pKa = 11.09DD171 pKa = 3.48NAISMRR177 pKa = 11.84NVFCQPRR184 pKa = 11.84IITTIMLEE192 pKa = 4.11KK193 pKa = 10.06EE194 pKa = 4.16VNEE197 pKa = 4.61AILWAKK203 pKa = 10.23RR204 pKa = 11.84KK205 pKa = 10.23RR206 pKa = 11.84EE207 pKa = 3.73VLYY210 pKa = 11.23VNNHH214 pKa = 5.99LYY216 pKa = 10.9KK217 pKa = 10.54LDD219 pKa = 3.38GLRR222 pKa = 11.84NQGVLAHH229 pKa = 6.62HH230 pKa = 7.05PKK232 pKa = 10.41ACEE235 pKa = 3.62ILLSGGQRR243 pKa = 11.84GCFEE247 pKa = 4.41VNLPSEE253 pKa = 4.46SLIHH257 pKa = 5.83AMKK260 pKa = 10.72RR261 pKa = 11.84EE262 pKa = 3.92LQTILNEE269 pKa = 4.16VIMFSVPPISYY280 pKa = 7.52DD281 pKa = 3.4TLLTGYY287 pKa = 10.31SKK289 pKa = 10.45ISRR292 pKa = 11.84IRR294 pKa = 11.84DD295 pKa = 3.34VPPIEE300 pKa = 4.11MRR302 pKa = 11.84LTALSALNRR311 pKa = 11.84LHH313 pKa = 7.77DD314 pKa = 4.06EE315 pKa = 4.14VTNTKK320 pKa = 8.7GTLFMTVYY328 pKa = 10.73ANKK331 pKa = 8.9GTGKK335 pKa = 7.53TTMTKK340 pKa = 10.02IMCEE344 pKa = 3.82KK345 pKa = 10.45LADD348 pKa = 3.4IYY350 pKa = 11.15KK351 pKa = 9.57RR352 pKa = 11.84PCYY355 pKa = 10.21RR356 pKa = 11.84IDD358 pKa = 3.3SDD360 pKa = 5.2AVGRR364 pKa = 11.84WLQTDD369 pKa = 3.3MSKK372 pKa = 10.49EE373 pKa = 3.95QFQKK377 pKa = 8.27MTFEE381 pKa = 5.08EE382 pKa = 3.93ILKK385 pKa = 10.06FNEE388 pKa = 4.12YY389 pKa = 8.69STSIYY394 pKa = 10.92EE395 pKa = 4.1VMAEE399 pKa = 4.13NIVAKK404 pKa = 10.72LGDD407 pKa = 3.5IKK409 pKa = 11.02LEE411 pKa = 3.92QILRR415 pKa = 11.84GRR417 pKa = 11.84YY418 pKa = 9.14SRR420 pKa = 11.84VNKK423 pKa = 10.27AIEE426 pKa = 3.81QFRR429 pKa = 11.84VEE431 pKa = 3.93RR432 pKa = 11.84GEE434 pKa = 4.36YY435 pKa = 10.04ILTTKK440 pKa = 10.5DD441 pKa = 3.34YY442 pKa = 9.91PAEE445 pKa = 3.66HH446 pKa = 6.63HH447 pKa = 6.97FYY449 pKa = 11.44SKK451 pKa = 10.22IASIAPYY458 pKa = 9.18GSIVIVEE465 pKa = 4.16GHH467 pKa = 6.03TLSQDD472 pKa = 2.95AYY474 pKa = 10.27LAPTNVSLIYY484 pKa = 10.21EE485 pKa = 4.67SIMDD489 pKa = 4.68PYY491 pKa = 11.21LAFNSRR497 pKa = 11.84DD498 pKa = 3.52NAKK501 pKa = 9.24TEE503 pKa = 3.92LFLYY507 pKa = 10.22LAYY510 pKa = 10.42DD511 pKa = 3.42RR512 pKa = 11.84EE513 pKa = 4.5LAHH516 pKa = 6.54SHH518 pKa = 5.94TKK520 pKa = 10.31FCMLEE525 pKa = 3.97FNDD528 pKa = 3.73WAVMRR533 pKa = 11.84SRR535 pKa = 11.84SNN537 pKa = 2.96
MM1 pKa = 7.7LFTINIRR8 pKa = 11.84APKK11 pKa = 9.14TLATLNRR18 pKa = 11.84MGIYY22 pKa = 10.24LRR24 pKa = 11.84EE25 pKa = 4.04QYY27 pKa = 10.64KK28 pKa = 10.42HH29 pKa = 6.13EE30 pKa = 4.28KK31 pKa = 8.72TSKK34 pKa = 10.77LLLKK38 pKa = 10.78RR39 pKa = 11.84NDD41 pKa = 4.13EE42 pKa = 4.02LHH44 pKa = 6.43HH45 pKa = 5.9WLEE48 pKa = 4.39KK49 pKa = 10.56LSARR53 pKa = 11.84HH54 pKa = 5.96KK55 pKa = 9.81GHH57 pKa = 6.14VFSMDD62 pKa = 2.66AVARR66 pKa = 11.84ILTEE70 pKa = 3.72RR71 pKa = 11.84LMKK74 pKa = 10.38INIIDD79 pKa = 3.57KK80 pKa = 10.71RR81 pKa = 11.84LGKK84 pKa = 10.74VKK86 pKa = 10.53DD87 pKa = 3.24IFANEE92 pKa = 4.38SKK94 pKa = 11.0NILQIVNKK102 pKa = 9.95EE103 pKa = 4.15DD104 pKa = 3.39VDD106 pKa = 4.47GVAMTYY112 pKa = 11.19DD113 pKa = 3.68DD114 pKa = 5.53ASFLLTLTKK123 pKa = 10.47EE124 pKa = 4.26VVIPNNEE131 pKa = 3.95TPVLYY136 pKa = 10.59EE137 pKa = 3.98IDD139 pKa = 4.19HH140 pKa = 6.12PTLRR144 pKa = 11.84HH145 pKa = 6.55LIPSYY150 pKa = 9.64IDD152 pKa = 3.04QMRR155 pKa = 11.84KK156 pKa = 8.42VLIIRR161 pKa = 11.84RR162 pKa = 11.84VTITDD167 pKa = 3.34VLYY170 pKa = 11.09DD171 pKa = 3.48NAISMRR177 pKa = 11.84NVFCQPRR184 pKa = 11.84IITTIMLEE192 pKa = 4.11KK193 pKa = 10.06EE194 pKa = 4.16VNEE197 pKa = 4.61AILWAKK203 pKa = 10.23RR204 pKa = 11.84KK205 pKa = 10.23RR206 pKa = 11.84EE207 pKa = 3.73VLYY210 pKa = 11.23VNNHH214 pKa = 5.99LYY216 pKa = 10.9KK217 pKa = 10.54LDD219 pKa = 3.38GLRR222 pKa = 11.84NQGVLAHH229 pKa = 6.62HH230 pKa = 7.05PKK232 pKa = 10.41ACEE235 pKa = 3.62ILLSGGQRR243 pKa = 11.84GCFEE247 pKa = 4.41VNLPSEE253 pKa = 4.46SLIHH257 pKa = 5.83AMKK260 pKa = 10.72RR261 pKa = 11.84EE262 pKa = 3.92LQTILNEE269 pKa = 4.16VIMFSVPPISYY280 pKa = 7.52DD281 pKa = 3.4TLLTGYY287 pKa = 10.31SKK289 pKa = 10.45ISRR292 pKa = 11.84IRR294 pKa = 11.84DD295 pKa = 3.34VPPIEE300 pKa = 4.11MRR302 pKa = 11.84LTALSALNRR311 pKa = 11.84LHH313 pKa = 7.77DD314 pKa = 4.06EE315 pKa = 4.14VTNTKK320 pKa = 8.7GTLFMTVYY328 pKa = 10.73ANKK331 pKa = 8.9GTGKK335 pKa = 7.53TTMTKK340 pKa = 10.02IMCEE344 pKa = 3.82KK345 pKa = 10.45LADD348 pKa = 3.4IYY350 pKa = 11.15KK351 pKa = 9.57RR352 pKa = 11.84PCYY355 pKa = 10.21RR356 pKa = 11.84IDD358 pKa = 3.3SDD360 pKa = 5.2AVGRR364 pKa = 11.84WLQTDD369 pKa = 3.3MSKK372 pKa = 10.49EE373 pKa = 3.95QFQKK377 pKa = 8.27MTFEE381 pKa = 5.08EE382 pKa = 3.93ILKK385 pKa = 10.06FNEE388 pKa = 4.12YY389 pKa = 8.69STSIYY394 pKa = 10.92EE395 pKa = 4.1VMAEE399 pKa = 4.13NIVAKK404 pKa = 10.72LGDD407 pKa = 3.5IKK409 pKa = 11.02LEE411 pKa = 3.92QILRR415 pKa = 11.84GRR417 pKa = 11.84YY418 pKa = 9.14SRR420 pKa = 11.84VNKK423 pKa = 10.27AIEE426 pKa = 3.81QFRR429 pKa = 11.84VEE431 pKa = 3.93RR432 pKa = 11.84GEE434 pKa = 4.36YY435 pKa = 10.04ILTTKK440 pKa = 10.5DD441 pKa = 3.34YY442 pKa = 9.91PAEE445 pKa = 3.66HH446 pKa = 6.63HH447 pKa = 6.97FYY449 pKa = 11.44SKK451 pKa = 10.22IASIAPYY458 pKa = 9.18GSIVIVEE465 pKa = 4.16GHH467 pKa = 6.03TLSQDD472 pKa = 2.95AYY474 pKa = 10.27LAPTNVSLIYY484 pKa = 10.21EE485 pKa = 4.67SIMDD489 pKa = 4.68PYY491 pKa = 11.21LAFNSRR497 pKa = 11.84DD498 pKa = 3.52NAKK501 pKa = 9.24TEE503 pKa = 3.92LFLYY507 pKa = 10.22LAYY510 pKa = 10.42DD511 pKa = 3.42RR512 pKa = 11.84EE513 pKa = 4.5LAHH516 pKa = 6.54SHH518 pKa = 5.94TKK520 pKa = 10.31FCMLEE525 pKa = 3.97FNDD528 pKa = 3.73WAVMRR533 pKa = 11.84SRR535 pKa = 11.84SNN537 pKa = 2.96
Molecular weight: 62.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5623 |
300 |
1285 |
702.9 |
80.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.438 ± 0.491 | 0.96 ± 0.23 |
5.691 ± 0.317 | 6.971 ± 0.278 |
3.575 ± 0.204 | 4.659 ± 0.469 |
2.294 ± 0.233 | 7.452 ± 0.383 |
4.802 ± 0.488 | 9.319 ± 0.429 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.934 ± 0.211 | 6.384 ± 0.353 |
3.254 ± 0.204 | 3.379 ± 0.375 |
6.029 ± 0.32 | 7.505 ± 0.493 |
6.758 ± 0.398 | 6.367 ± 0.254 |
1.014 ± 0.125 | 4.215 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
