
Prochlorococcus marinus (strain MIT 9211)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Prochloraceae; Prochlorococcus; Prochlorococcus marinus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
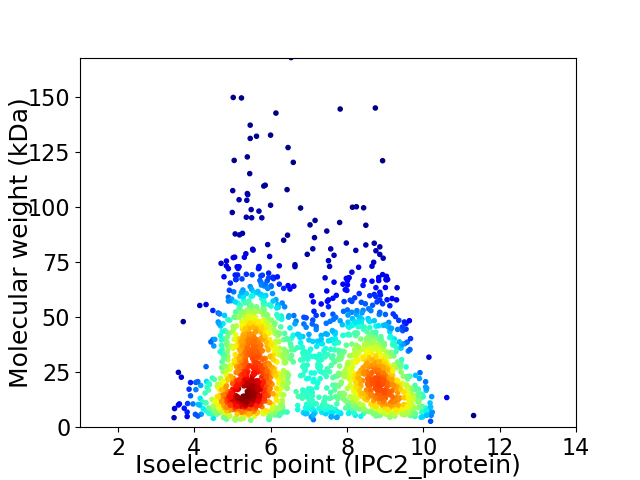
Virtual 2D-PAGE plot for 1855 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A9BAA0|A9BAA0_PROM4 Uncharacterized protein OS=Prochlorococcus marinus (strain MIT 9211) OX=93059 GN=P9211_08311 PE=4 SV=1
MM1 pKa = 7.72KK2 pKa = 10.51LFQRR6 pKa = 11.84LLVAPAALGLMAPLAANAAEE26 pKa = 4.5VNIQDD31 pKa = 3.59VGNYY35 pKa = 10.06AGLEE39 pKa = 4.08EE40 pKa = 4.21QHH42 pKa = 6.69VEE44 pKa = 4.16TTAQFSDD51 pKa = 4.06VVPGDD56 pKa = 3.31WAYY59 pKa = 8.91TALQNLSEE67 pKa = 4.63SYY69 pKa = 10.7GCVGNAYY76 pKa = 10.01SQNLKK81 pKa = 10.37SGQALTRR88 pKa = 11.84YY89 pKa = 7.99EE90 pKa = 4.12AASLINSCLEE100 pKa = 4.13GGLVASGQGLSQDD113 pKa = 3.44AARR116 pKa = 11.84LSNEE120 pKa = 4.47FGSEE124 pKa = 3.69MAILKK129 pKa = 10.25GRR131 pKa = 11.84VDD133 pKa = 3.41GLEE136 pKa = 3.74YY137 pKa = 10.8RR138 pKa = 11.84LNEE141 pKa = 3.99VNAGTFANTTLISGSATFTVGAVDD165 pKa = 5.09DD166 pKa = 4.22STSGEE171 pKa = 4.2KK172 pKa = 10.21LHH174 pKa = 7.14SIYY177 pKa = 10.58AYY179 pKa = 10.03GIDD182 pKa = 3.54MSTSFSGEE190 pKa = 3.85DD191 pKa = 3.43SLDD194 pKa = 3.47VAVIAGNAGTQPLNVLDD211 pKa = 4.12SAEE214 pKa = 4.07GTTNQLQIDD223 pKa = 4.07KK224 pKa = 8.69LTYY227 pKa = 9.97SFPVGDD233 pKa = 4.2FRR235 pKa = 11.84VTAGPLMDD243 pKa = 3.94QDD245 pKa = 5.43DD246 pKa = 4.82IISAKK251 pKa = 10.15LSSYY255 pKa = 10.41SAEE258 pKa = 4.37FYY260 pKa = 8.95ITPHH264 pKa = 6.2EE265 pKa = 4.15YY266 pKa = 10.56SRR268 pKa = 11.84AEE270 pKa = 4.11TEE272 pKa = 3.91EE273 pKa = 4.74GPGVGATYY281 pKa = 10.37FFDD284 pKa = 4.29NKK286 pKa = 8.99WNASISAVFDD296 pKa = 4.78DD297 pKa = 6.24GDD299 pKa = 3.92DD300 pKa = 3.64ASNGILTRR308 pKa = 11.84EE309 pKa = 4.1GDD311 pKa = 3.92DD312 pKa = 4.26VITGSIGYY320 pKa = 9.92DD321 pKa = 3.12GDD323 pKa = 3.43GWGGGVILSSGDD335 pKa = 3.44KK336 pKa = 10.68NGSDD340 pKa = 3.38TTDD343 pKa = 3.24YY344 pKa = 11.59DD345 pKa = 3.39SWGWGLYY352 pKa = 8.75WEE354 pKa = 5.26PEE356 pKa = 3.91DD357 pKa = 4.93FPRR360 pKa = 11.84FSAGYY365 pKa = 7.65DD366 pKa = 3.32TQEE369 pKa = 4.23NEE371 pKa = 3.84GAADD375 pKa = 3.54NDD377 pKa = 3.27SWYY380 pKa = 10.14IAADD384 pKa = 3.55YY385 pKa = 10.76DD386 pKa = 3.69GWGPGTLSAAYY397 pKa = 9.36QVIDD401 pKa = 4.75DD402 pKa = 4.46EE403 pKa = 4.83TNEE406 pKa = 3.8LAAFEE411 pKa = 5.29VYY413 pKa = 10.78YY414 pKa = 10.89NWDD417 pKa = 3.43VADD420 pKa = 5.29GISVRR425 pKa = 11.84PGLFTQEE432 pKa = 3.96VAGGEE437 pKa = 4.3DD438 pKa = 3.12EE439 pKa = 4.34TGAVVEE445 pKa = 4.7TTFKK449 pKa = 10.93FF450 pKa = 3.57
MM1 pKa = 7.72KK2 pKa = 10.51LFQRR6 pKa = 11.84LLVAPAALGLMAPLAANAAEE26 pKa = 4.5VNIQDD31 pKa = 3.59VGNYY35 pKa = 10.06AGLEE39 pKa = 4.08EE40 pKa = 4.21QHH42 pKa = 6.69VEE44 pKa = 4.16TTAQFSDD51 pKa = 4.06VVPGDD56 pKa = 3.31WAYY59 pKa = 8.91TALQNLSEE67 pKa = 4.63SYY69 pKa = 10.7GCVGNAYY76 pKa = 10.01SQNLKK81 pKa = 10.37SGQALTRR88 pKa = 11.84YY89 pKa = 7.99EE90 pKa = 4.12AASLINSCLEE100 pKa = 4.13GGLVASGQGLSQDD113 pKa = 3.44AARR116 pKa = 11.84LSNEE120 pKa = 4.47FGSEE124 pKa = 3.69MAILKK129 pKa = 10.25GRR131 pKa = 11.84VDD133 pKa = 3.41GLEE136 pKa = 3.74YY137 pKa = 10.8RR138 pKa = 11.84LNEE141 pKa = 3.99VNAGTFANTTLISGSATFTVGAVDD165 pKa = 5.09DD166 pKa = 4.22STSGEE171 pKa = 4.2KK172 pKa = 10.21LHH174 pKa = 7.14SIYY177 pKa = 10.58AYY179 pKa = 10.03GIDD182 pKa = 3.54MSTSFSGEE190 pKa = 3.85DD191 pKa = 3.43SLDD194 pKa = 3.47VAVIAGNAGTQPLNVLDD211 pKa = 4.12SAEE214 pKa = 4.07GTTNQLQIDD223 pKa = 4.07KK224 pKa = 8.69LTYY227 pKa = 9.97SFPVGDD233 pKa = 4.2FRR235 pKa = 11.84VTAGPLMDD243 pKa = 3.94QDD245 pKa = 5.43DD246 pKa = 4.82IISAKK251 pKa = 10.15LSSYY255 pKa = 10.41SAEE258 pKa = 4.37FYY260 pKa = 8.95ITPHH264 pKa = 6.2EE265 pKa = 4.15YY266 pKa = 10.56SRR268 pKa = 11.84AEE270 pKa = 4.11TEE272 pKa = 3.91EE273 pKa = 4.74GPGVGATYY281 pKa = 10.37FFDD284 pKa = 4.29NKK286 pKa = 8.99WNASISAVFDD296 pKa = 4.78DD297 pKa = 6.24GDD299 pKa = 3.92DD300 pKa = 3.64ASNGILTRR308 pKa = 11.84EE309 pKa = 4.1GDD311 pKa = 3.92DD312 pKa = 4.26VITGSIGYY320 pKa = 9.92DD321 pKa = 3.12GDD323 pKa = 3.43GWGGGVILSSGDD335 pKa = 3.44KK336 pKa = 10.68NGSDD340 pKa = 3.38TTDD343 pKa = 3.24YY344 pKa = 11.59DD345 pKa = 3.39SWGWGLYY352 pKa = 8.75WEE354 pKa = 5.26PEE356 pKa = 3.91DD357 pKa = 4.93FPRR360 pKa = 11.84FSAGYY365 pKa = 7.65DD366 pKa = 3.32TQEE369 pKa = 4.23NEE371 pKa = 3.84GAADD375 pKa = 3.54NDD377 pKa = 3.27SWYY380 pKa = 10.14IAADD384 pKa = 3.55YY385 pKa = 10.76DD386 pKa = 3.69GWGPGTLSAAYY397 pKa = 9.36QVIDD401 pKa = 4.75DD402 pKa = 4.46EE403 pKa = 4.83TNEE406 pKa = 3.8LAAFEE411 pKa = 5.29VYY413 pKa = 10.78YY414 pKa = 10.89NWDD417 pKa = 3.43VADD420 pKa = 5.29GISVRR425 pKa = 11.84PGLFTQEE432 pKa = 3.96VAGGEE437 pKa = 4.3DD438 pKa = 3.12EE439 pKa = 4.34TGAVVEE445 pKa = 4.7TTFKK449 pKa = 10.93FF450 pKa = 3.57
Molecular weight: 47.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A9BBJ0|RNPA_PROM4 Ribonuclease P protein component OS=Prochlorococcus marinus (strain MIT 9211) OX=93059 GN=rnpA PE=3 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84THH26 pKa = 5.95TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.38KK38 pKa = 10.52GRR40 pKa = 11.84SQLAVV45 pKa = 3.09
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.07RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84THH26 pKa = 5.95TGRR29 pKa = 11.84RR30 pKa = 11.84VIRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.38KK38 pKa = 10.52GRR40 pKa = 11.84SQLAVV45 pKa = 3.09
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
505001 |
25 |
1531 |
272.2 |
30.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.16 ± 0.079 | 1.214 ± 0.021 |
4.966 ± 0.053 | 6.406 ± 0.064 |
3.987 ± 0.045 | 7.053 ± 0.053 |
1.843 ± 0.028 | 7.45 ± 0.058 |
6.305 ± 0.067 | 11.673 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.978 ± 0.026 | 4.684 ± 0.042 |
4.372 ± 0.035 | 3.715 ± 0.036 |
5.105 ± 0.046 | 7.681 ± 0.042 |
4.685 ± 0.039 | 5.839 ± 0.05 |
1.472 ± 0.031 | 2.411 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
