
Phocoena spinipinnis papillomavirus (isolate Burmeister s porpoise/Peru/PsPV1) (PsPV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Omikronpapillomavirus; Omikronpapillomavirus 1
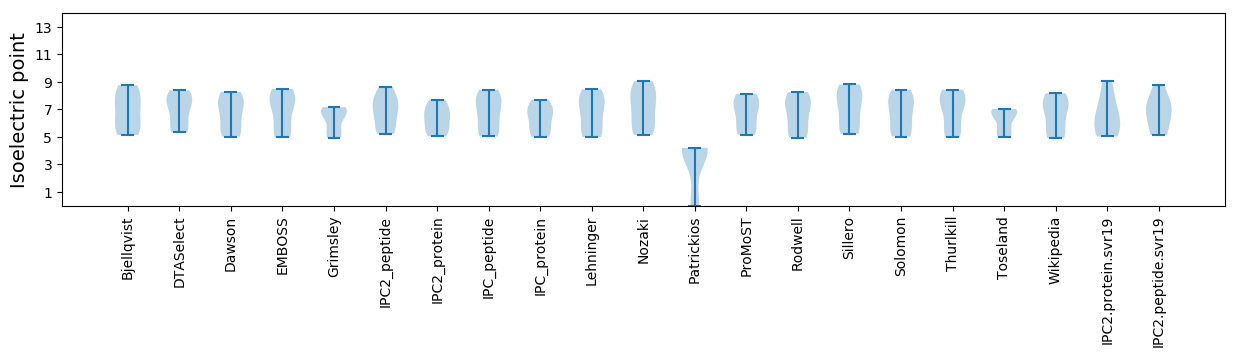
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
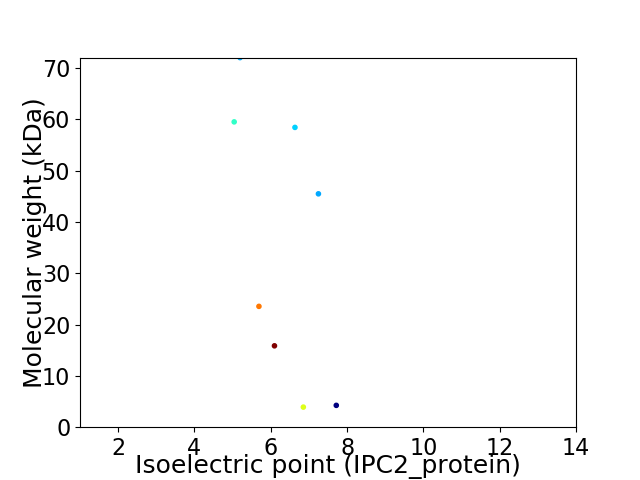
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8UZ14|Q8UZ14_PSPVP E5b protein (Fragment) OS=Phocoena spinipinnis papillomavirus (isolate Burmeister's porpoise/Peru/PsPV1) OX=654916 GN=e5b PE=4 SV=1
MM1 pKa = 6.98VRR3 pKa = 11.84VKK5 pKa = 10.36RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AAEE12 pKa = 3.75GDD14 pKa = 3.74LYY16 pKa = 11.14AGCRR20 pKa = 11.84RR21 pKa = 11.84GQDD24 pKa = 3.42CPDD27 pKa = 4.52DD28 pKa = 3.73IKK30 pKa = 11.33PKK32 pKa = 10.54FEE34 pKa = 3.81QDD36 pKa = 2.38TWADD40 pKa = 3.89RR41 pKa = 11.84FLKK44 pKa = 9.65WFSSIIYY51 pKa = 9.6LGNLGISTGRR61 pKa = 11.84GAGGSTGYY69 pKa = 10.76VPVGSGGGRR78 pKa = 11.84GVRR81 pKa = 11.84PAMGGQPSRR90 pKa = 11.84PNVVVEE96 pKa = 4.1NVGPAEE102 pKa = 4.18VPVDD106 pKa = 3.75GAVDD110 pKa = 3.9ASAPSVITPSEE121 pKa = 4.16STVVVGGSTTPHH133 pKa = 6.33EE134 pKa = 5.15EE135 pKa = 3.85IPLVPLHH142 pKa = 6.78PEE144 pKa = 3.81VGPDD148 pKa = 3.84PEE150 pKa = 6.17PGLPLPPPEE159 pKa = 5.21SGGPAVLDD167 pKa = 3.24VTLNVTSTYY176 pKa = 9.5THH178 pKa = 7.39DD179 pKa = 3.91PSIIHH184 pKa = 6.36PRR186 pKa = 11.84VSSLGEE192 pKa = 3.9SAGAEE197 pKa = 3.69APLFPTISLQPLDD210 pKa = 4.04VSLLPGEE217 pKa = 5.33SSFQPHH223 pKa = 6.97AYY225 pKa = 8.98IDD227 pKa = 3.73LSGSFEE233 pKa = 4.45EE234 pKa = 4.75IEE236 pKa = 4.23LDD238 pKa = 3.55AFTSGPQDD246 pKa = 3.72PQTSTPMSRR255 pKa = 11.84VDD257 pKa = 3.0SGLRR261 pKa = 11.84SVRR264 pKa = 11.84RR265 pKa = 11.84AYY267 pKa = 10.47SRR269 pKa = 11.84RR270 pKa = 11.84TGALRR275 pKa = 11.84RR276 pKa = 11.84LYY278 pKa = 10.46HH279 pKa = 7.34RR280 pKa = 11.84LTQQVRR286 pKa = 11.84VNRR289 pKa = 11.84PEE291 pKa = 3.7FLRR294 pKa = 11.84RR295 pKa = 11.84PSQLVSYY302 pKa = 10.5VFDD305 pKa = 3.83NAAFDD310 pKa = 4.75PDD312 pKa = 3.97TTLHH316 pKa = 6.61FPQASEE322 pKa = 5.2DD323 pKa = 3.8VLQAPDD329 pKa = 5.37LDD331 pKa = 4.15FQDD334 pKa = 3.7VGTLHH339 pKa = 6.77RR340 pKa = 11.84PIYY343 pKa = 8.41STEE346 pKa = 3.64GGYY349 pKa = 10.54VRR351 pKa = 11.84VSRR354 pKa = 11.84FGEE357 pKa = 3.95RR358 pKa = 11.84EE359 pKa = 3.98TIRR362 pKa = 11.84TRR364 pKa = 11.84SGAAIGARR372 pKa = 11.84VHH374 pKa = 7.1FYY376 pKa = 10.8TDD378 pKa = 3.65LSSIQSFAEE387 pKa = 3.74QLPSVGSLGPDD398 pKa = 3.06VADD401 pKa = 4.55PGIEE405 pKa = 3.61LHH407 pKa = 6.88LFGEE411 pKa = 5.1GTGDD415 pKa = 3.37TSIADD420 pKa = 3.64AQGGGVSLSNGTLHH434 pKa = 6.87TEE436 pKa = 4.24TEE438 pKa = 4.22FTNASNGSLHH448 pKa = 6.81SEE450 pKa = 4.32YY451 pKa = 10.77SNSMLLDD458 pKa = 3.9SYY460 pKa = 11.63TEE462 pKa = 4.22TFNDD466 pKa = 3.41AQLALMDD473 pKa = 4.73SEE475 pKa = 5.5GSTQVLSIPEE485 pKa = 4.02LARR488 pKa = 11.84PVRR491 pKa = 11.84GFAEE495 pKa = 4.47STGGLSVSYY504 pKa = 9.51PVDD507 pKa = 3.5MEE509 pKa = 4.14ISGSSTSFIHH519 pKa = 6.8NIPGPPSILLFYY531 pKa = 9.82PDD533 pKa = 4.64SSPSFYY539 pKa = 10.86LHH541 pKa = 7.07PSLLRR546 pKa = 11.84RR547 pKa = 11.84KK548 pKa = 9.49RR549 pKa = 11.84KK550 pKa = 9.18RR551 pKa = 11.84VFYY554 pKa = 10.92
MM1 pKa = 6.98VRR3 pKa = 11.84VKK5 pKa = 10.36RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AAEE12 pKa = 3.75GDD14 pKa = 3.74LYY16 pKa = 11.14AGCRR20 pKa = 11.84RR21 pKa = 11.84GQDD24 pKa = 3.42CPDD27 pKa = 4.52DD28 pKa = 3.73IKK30 pKa = 11.33PKK32 pKa = 10.54FEE34 pKa = 3.81QDD36 pKa = 2.38TWADD40 pKa = 3.89RR41 pKa = 11.84FLKK44 pKa = 9.65WFSSIIYY51 pKa = 9.6LGNLGISTGRR61 pKa = 11.84GAGGSTGYY69 pKa = 10.76VPVGSGGGRR78 pKa = 11.84GVRR81 pKa = 11.84PAMGGQPSRR90 pKa = 11.84PNVVVEE96 pKa = 4.1NVGPAEE102 pKa = 4.18VPVDD106 pKa = 3.75GAVDD110 pKa = 3.9ASAPSVITPSEE121 pKa = 4.16STVVVGGSTTPHH133 pKa = 6.33EE134 pKa = 5.15EE135 pKa = 3.85IPLVPLHH142 pKa = 6.78PEE144 pKa = 3.81VGPDD148 pKa = 3.84PEE150 pKa = 6.17PGLPLPPPEE159 pKa = 5.21SGGPAVLDD167 pKa = 3.24VTLNVTSTYY176 pKa = 9.5THH178 pKa = 7.39DD179 pKa = 3.91PSIIHH184 pKa = 6.36PRR186 pKa = 11.84VSSLGEE192 pKa = 3.9SAGAEE197 pKa = 3.69APLFPTISLQPLDD210 pKa = 4.04VSLLPGEE217 pKa = 5.33SSFQPHH223 pKa = 6.97AYY225 pKa = 8.98IDD227 pKa = 3.73LSGSFEE233 pKa = 4.45EE234 pKa = 4.75IEE236 pKa = 4.23LDD238 pKa = 3.55AFTSGPQDD246 pKa = 3.72PQTSTPMSRR255 pKa = 11.84VDD257 pKa = 3.0SGLRR261 pKa = 11.84SVRR264 pKa = 11.84RR265 pKa = 11.84AYY267 pKa = 10.47SRR269 pKa = 11.84RR270 pKa = 11.84TGALRR275 pKa = 11.84RR276 pKa = 11.84LYY278 pKa = 10.46HH279 pKa = 7.34RR280 pKa = 11.84LTQQVRR286 pKa = 11.84VNRR289 pKa = 11.84PEE291 pKa = 3.7FLRR294 pKa = 11.84RR295 pKa = 11.84PSQLVSYY302 pKa = 10.5VFDD305 pKa = 3.83NAAFDD310 pKa = 4.75PDD312 pKa = 3.97TTLHH316 pKa = 6.61FPQASEE322 pKa = 5.2DD323 pKa = 3.8VLQAPDD329 pKa = 5.37LDD331 pKa = 4.15FQDD334 pKa = 3.7VGTLHH339 pKa = 6.77RR340 pKa = 11.84PIYY343 pKa = 8.41STEE346 pKa = 3.64GGYY349 pKa = 10.54VRR351 pKa = 11.84VSRR354 pKa = 11.84FGEE357 pKa = 3.95RR358 pKa = 11.84EE359 pKa = 3.98TIRR362 pKa = 11.84TRR364 pKa = 11.84SGAAIGARR372 pKa = 11.84VHH374 pKa = 7.1FYY376 pKa = 10.8TDD378 pKa = 3.65LSSIQSFAEE387 pKa = 3.74QLPSVGSLGPDD398 pKa = 3.06VADD401 pKa = 4.55PGIEE405 pKa = 3.61LHH407 pKa = 6.88LFGEE411 pKa = 5.1GTGDD415 pKa = 3.37TSIADD420 pKa = 3.64AQGGGVSLSNGTLHH434 pKa = 6.87TEE436 pKa = 4.24TEE438 pKa = 4.22FTNASNGSLHH448 pKa = 6.81SEE450 pKa = 4.32YY451 pKa = 10.77SNSMLLDD458 pKa = 3.9SYY460 pKa = 11.63TEE462 pKa = 4.22TFNDD466 pKa = 3.41AQLALMDD473 pKa = 4.73SEE475 pKa = 5.5GSTQVLSIPEE485 pKa = 4.02LARR488 pKa = 11.84PVRR491 pKa = 11.84GFAEE495 pKa = 4.47STGGLSVSYY504 pKa = 9.51PVDD507 pKa = 3.5MEE509 pKa = 4.14ISGSSTSFIHH519 pKa = 6.8NIPGPPSILLFYY531 pKa = 9.82PDD533 pKa = 4.64SSPSFYY539 pKa = 10.86LHH541 pKa = 7.07PSLLRR546 pKa = 11.84RR547 pKa = 11.84KK548 pKa = 9.49RR549 pKa = 11.84KK550 pKa = 9.18RR551 pKa = 11.84VFYY554 pKa = 10.92
Molecular weight: 59.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8UZ16|Q8UZ16_PSPVP E4 protein (Fragment) OS=Phocoena spinipinnis papillomavirus (isolate Burmeister's porpoise/Peru/PsPV1) OX=654916 GN=e4 PE=4 SV=1
TT1 pKa = 7.19AQRR4 pKa = 11.84CSCWQRR10 pKa = 11.84KK11 pKa = 7.39SAFLALFSFVCICAYY26 pKa = 9.64LCTYY30 pKa = 9.55ISLYY34 pKa = 9.98IALL37 pKa = 4.57
TT1 pKa = 7.19AQRR4 pKa = 11.84CSCWQRR10 pKa = 11.84KK11 pKa = 7.39SAFLALFSFVCICAYY26 pKa = 9.64LCTYY30 pKa = 9.55ISLYY34 pKa = 9.98IALL37 pKa = 4.57
Molecular weight: 4.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2558 |
34 |
642 |
319.8 |
35.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.646 ± 0.571 | 2.658 ± 0.772 |
5.786 ± 0.629 | 5.786 ± 0.403 |
3.753 ± 0.484 | 7.193 ± 0.814 |
2.737 ± 0.64 | 3.714 ± 0.386 |
4.457 ± 0.962 | 8.835 ± 0.842 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.798 ± 0.336 | 3.245 ± 0.519 |
7.545 ± 0.984 | 3.909 ± 0.398 |
5.708 ± 0.396 | 9.03 ± 0.927 |
6.255 ± 0.311 | 6.216 ± 0.425 |
1.642 ± 0.348 | 3.088 ± 0.308 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
