
Lactobacillus thailandensis DSM 22698 = JCM 13996
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lacticaseibacillus; Lacticaseibacillus thailandensis
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
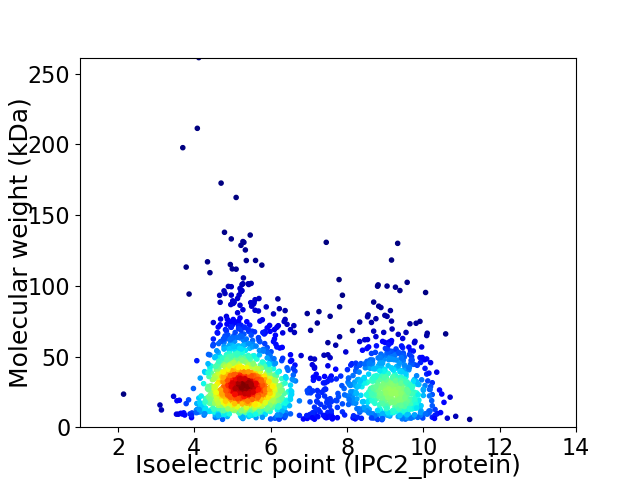
Virtual 2D-PAGE plot for 1860 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R2C7R1|A0A0R2C7R1_9LACO C-di-GMP-specific phosphodiesterase OS=Lactobacillus thailandensis DSM 22698 = JCM 13996 OX=1423810 GN=FD19_GL001104 PE=4 SV=1
MM1 pKa = 7.95LSPQVQNVAASTATAGSTTEE21 pKa = 4.04EE22 pKa = 4.05TSGITNTSVAHH33 pKa = 6.36SSADD37 pKa = 3.34VKK39 pKa = 11.3ASVASTSSATVVNTGNDD56 pKa = 3.17SSTATSATSTASASTSVSAVASATNGSAEE85 pKa = 4.17SLAATSAATAATTSVAVDD103 pKa = 3.94DD104 pKa = 4.93SSAALPASSAATAGAEE120 pKa = 4.29TSSATSGSSAASAQTVTSVAHH141 pKa = 4.67QALRR145 pKa = 11.84TRR147 pKa = 11.84SLASSAAPADD157 pKa = 4.05SEE159 pKa = 4.48VTTAINSGAANAPATVSYY177 pKa = 10.87HH178 pKa = 6.11NGSTQPTISVMVSGNFQAGDD198 pKa = 3.66VVTVYY203 pKa = 11.26LDD205 pKa = 3.49TTAMSVTAATTVKK218 pKa = 10.4GATEE222 pKa = 4.3SNVDD226 pKa = 2.93VSTNDD231 pKa = 2.63GRR233 pKa = 11.84TVKK236 pKa = 10.26GAQYY240 pKa = 10.61QFSSDD245 pKa = 3.57YY246 pKa = 10.94NGAFPVVFVLNGTADD261 pKa = 3.65DD262 pKa = 4.34QINNGTSTATIAVDD276 pKa = 3.22VDD278 pKa = 3.77KK279 pKa = 11.5NGTIVATNSVVLDD292 pKa = 3.58KK293 pKa = 10.82SYY295 pKa = 10.48TMTQPNDD302 pKa = 3.62TIRR305 pKa = 11.84RR306 pKa = 11.84NILNGTNGVTAGVIAQGAVTSEE328 pKa = 3.92LTLRR332 pKa = 11.84AQLQGLQYY340 pKa = 10.69FGDD343 pKa = 3.94YY344 pKa = 8.83STLVNSSPYY353 pKa = 10.35EE354 pKa = 4.0KK355 pKa = 9.79LTAASTVTFDD365 pKa = 6.0FSDD368 pKa = 3.51QLKK371 pKa = 10.21VDD373 pKa = 4.02SAVLSDD379 pKa = 4.17DD380 pKa = 3.84AVATGVTLTRR390 pKa = 11.84SGNSVTLTLPSGLTLAAISKK410 pKa = 9.8LAGYY414 pKa = 8.69LQVGLLPVTVPTTSTRR430 pKa = 11.84YY431 pKa = 9.63VLPTLTATWTIDD443 pKa = 3.13GQTNTVTEE451 pKa = 4.51PGNSGSSHH459 pKa = 6.3ANEE462 pKa = 3.7FAYY465 pKa = 10.64NIQTYY470 pKa = 7.62TAVGTVTVSSSSSYY484 pKa = 11.08LGLLQQGTDD493 pKa = 3.19NDD495 pKa = 4.39YY496 pKa = 10.83GLQWVVANEE505 pKa = 4.32SNFASGTTTTGSLSITSSPDD525 pKa = 2.86NSVGITSVNGLDD537 pKa = 3.88GNYY540 pKa = 7.86LTSAGLSKK548 pKa = 10.96NLTITFTTADD558 pKa = 3.63GQSQSFEE565 pKa = 4.31YY566 pKa = 11.11VDD568 pKa = 3.6GTTSYY573 pKa = 11.08VATVGSATDD582 pKa = 3.68PVTQVQIVDD591 pKa = 4.04HH592 pKa = 6.77AAQTAPYY599 pKa = 7.97VTGSISHH606 pKa = 6.1LQVGLIAYY614 pKa = 9.29GDD616 pKa = 3.77EE617 pKa = 4.31KK618 pKa = 9.81TTVTATADD626 pKa = 3.17TSLYY630 pKa = 11.0DD631 pKa = 3.67GDD633 pKa = 4.31GKK635 pKa = 11.11LVLDD639 pKa = 4.43TASSTDD645 pKa = 3.02SDD647 pKa = 4.14VVRR650 pKa = 11.84EE651 pKa = 4.01RR652 pKa = 11.84TVTKK656 pKa = 9.67TYY658 pKa = 10.42YY659 pKa = 8.19PQPSIVLSAGSNGVDD674 pKa = 3.01NQQGNTAQQIYY685 pKa = 10.71NPGDD689 pKa = 3.55EE690 pKa = 4.4LTEE693 pKa = 4.31SVNLLDD699 pKa = 4.95GGTNSGSASDD709 pKa = 3.52QVGRR713 pKa = 11.84IGNGDD718 pKa = 3.51GKK720 pKa = 10.56QPAVEE725 pKa = 4.67AVAAQNPMTIVVVAPTNTDD744 pKa = 3.49FADD747 pKa = 4.09DD748 pKa = 3.76ATLLSTLNKK757 pKa = 10.16SDD759 pKa = 3.33ISSSGDD765 pKa = 3.41LEE767 pKa = 4.2ITHH770 pKa = 7.15LDD772 pKa = 3.82DD773 pKa = 3.65VDD775 pKa = 3.81GRR777 pKa = 11.84PVIAITGVDD786 pKa = 3.67HH787 pKa = 5.71YY788 pKa = 9.22TASSRR793 pKa = 11.84LPIVLQVNADD803 pKa = 4.0AIGGSTISALNTIYY817 pKa = 11.2VMPSAVLSQYY827 pKa = 11.71NNGTILTSDD836 pKa = 3.59GSLATDD842 pKa = 3.72WATGNMYY849 pKa = 10.68NLSTGGSQLGTIITASSFTANASITGSADD878 pKa = 2.58SSAQVQTGTINVDD891 pKa = 3.03DD892 pKa = 4.83DD893 pKa = 3.67QATGKK898 pKa = 6.27TTLYY902 pKa = 10.86NGTTNPVTDD911 pKa = 3.41TLTTVNLVDD920 pKa = 4.47ADD922 pKa = 3.81GNQYY926 pKa = 8.16ITLTGPVQVVDD937 pKa = 3.9DD938 pKa = 4.8AGNTVAAAVTYY949 pKa = 7.38YY950 pKa = 11.03QNGAVVTDD958 pKa = 3.93PTQADD963 pKa = 3.97SFTVSGVSLAKK974 pKa = 9.97KK975 pKa = 10.1GSYY978 pKa = 10.44AVISYY983 pKa = 10.24GLTVVPSQLAGALTNKK999 pKa = 7.7TWAYY1003 pKa = 6.81QTSTQGYY1010 pKa = 5.64TAGTVATGNLVPLGAAISNTLVLQAIRR1037 pKa = 11.84DD1038 pKa = 4.02LTDD1041 pKa = 2.43TWYY1044 pKa = 10.33VAQRR1048 pKa = 11.84KK1049 pKa = 9.29ADD1051 pKa = 3.46GTYY1054 pKa = 10.46VVDD1057 pKa = 3.76SAVAAVTTTGVVGRR1071 pKa = 11.84IYY1073 pKa = 10.86NGYY1076 pKa = 7.5TLMGTSAADD1085 pKa = 3.1VGQAYY1090 pKa = 10.28GDD1092 pKa = 4.15DD1093 pKa = 3.88GAVLVGVYY1101 pKa = 10.29DD1102 pKa = 3.6ATTGQMVADD1111 pKa = 4.65GSMITYY1117 pKa = 10.32AEE1119 pKa = 4.11QVNSDD1124 pKa = 3.57GTITGHH1130 pKa = 5.76TYY1132 pKa = 11.26YY1133 pKa = 10.95LVTEE1137 pKa = 4.12QDD1139 pKa = 3.42VATKK1143 pKa = 10.79LNDD1146 pKa = 3.13VTVTYY1151 pKa = 10.55NGQTAASEE1159 pKa = 4.47VAGLQATANKK1169 pKa = 9.77VNVGLTSDD1177 pKa = 5.21DD1178 pKa = 3.21ITVVKK1183 pKa = 10.7DD1184 pKa = 3.74GINAGTYY1191 pKa = 8.91TYY1193 pKa = 11.03QLNAQGLAKK1202 pKa = 10.37LQAAVGNDD1210 pKa = 3.83AIVTDD1215 pKa = 3.92VGVTGTITIQPAVTTVTLNPGALTYY1240 pKa = 10.62DD1241 pKa = 3.14GRR1243 pKa = 11.84TPASGASLTATVAGQTLKK1261 pKa = 11.06LNSGDD1266 pKa = 3.44VTVADD1271 pKa = 4.78DD1272 pKa = 5.08GIDD1275 pKa = 3.35VGTYY1279 pKa = 9.01TYY1281 pKa = 11.18QLTAAGLSSLQSQVGSNYY1299 pKa = 9.79VVTTGTPGMIVISPEE1314 pKa = 3.83QMPVTLSSGSLVYY1327 pKa = 10.42DD1328 pKa = 4.38GKK1330 pKa = 9.4TKK1332 pKa = 10.87ASGAAVIATVNGEE1345 pKa = 4.31PLTLNDD1351 pKa = 5.4DD1352 pKa = 5.25DD1353 pKa = 4.2ITVTNDD1359 pKa = 3.87GINVGTYY1366 pKa = 10.1NYY1368 pKa = 10.43QLNQTGLEE1376 pKa = 4.03KK1377 pKa = 10.83LQTLVGDD1384 pKa = 4.13DD1385 pKa = 3.66FTVVVGDD1392 pKa = 4.47PGTITITPAVVDD1404 pKa = 3.71VVLNDD1409 pKa = 3.82GSFTYY1414 pKa = 10.46DD1415 pKa = 2.78GNTAASAAKK1424 pKa = 10.42GLVATVNGTPVDD1436 pKa = 4.38LDD1438 pKa = 3.54NSDD1441 pKa = 4.72IEE1443 pKa = 4.47VTDD1446 pKa = 4.59DD1447 pKa = 4.67GVDD1450 pKa = 3.22AGNYY1454 pKa = 9.6AYY1456 pKa = 10.49QLNDD1460 pKa = 3.17AGLAKK1465 pKa = 10.29VQLVVGSNYY1474 pKa = 9.37QVSVAVTGTITITPASITAALSDD1497 pKa = 4.11GGFTYY1502 pKa = 10.69DD1503 pKa = 3.36GATKK1507 pKa = 10.35ASEE1510 pKa = 4.28ATGLTATANSQAVDD1524 pKa = 3.71LSGEE1528 pKa = 4.42DD1529 pKa = 3.61IEE1531 pKa = 4.63VTNDD1535 pKa = 2.79GTGAGNYY1542 pKa = 9.65AYY1544 pKa = 10.13QLSDD1548 pKa = 3.09AGLAKK1553 pKa = 10.06VQSVVGTNYY1562 pKa = 9.75TVVLGGSGTVTIAQVVATTVXQSAVGPNYY1591 pKa = 9.78TVGVGDD1597 pKa = 4.13TGTITITPASITTTLSDD1614 pKa = 4.17GGFTYY1619 pKa = 10.71DD1620 pKa = 3.48GTTKK1624 pKa = 10.93ASGAPKK1630 pKa = 9.24LTATVVGQTVEE1641 pKa = 4.48LDD1643 pKa = 3.52SSDD1646 pKa = 4.41VDD1648 pKa = 3.8VTDD1651 pKa = 3.89NGSAAGDD1658 pKa = 3.44YY1659 pKa = 10.62VYY1661 pKa = 10.92QLNANGLEE1669 pKa = 4.38KK1670 pKa = 10.81VQSAAGANYY1679 pKa = 9.17IVRR1682 pKa = 11.84VGDD1685 pKa = 3.34TGTITIAKK1693 pKa = 9.71APATTTLSDD1702 pKa = 3.74GGFTYY1707 pKa = 10.71DD1708 pKa = 3.48GTTKK1712 pKa = 10.93ASGAPKK1718 pKa = 9.24LTATVVGQTVEE1729 pKa = 4.67LDD1731 pKa = 3.4SSDD1734 pKa = 3.09IDD1736 pKa = 4.18VINDD1740 pKa = 4.49AIAVGSYY1747 pKa = 10.35DD1748 pKa = 3.6YY1749 pKa = 11.14QLNATGLAKK1758 pKa = 10.47VQSAAGGNYY1767 pKa = 8.62TVGFGDD1773 pKa = 4.4AGTITITPASITTTLSDD1790 pKa = 4.17GGFTYY1795 pKa = 10.71DD1796 pKa = 3.48GTTKK1800 pKa = 10.93ASGAPKK1806 pKa = 9.34XFGDD1810 pKa = 4.38AGTITITPASITTTLSDD1827 pKa = 4.17GGFTYY1832 pKa = 10.71DD1833 pKa = 3.48GTTKK1837 pKa = 10.93ASGAPKK1843 pKa = 9.25LTATVIGQTVDD1854 pKa = 4.05LDD1856 pKa = 3.86SSDD1859 pKa = 4.12IDD1861 pKa = 3.52ITNDD1865 pKa = 2.45ATTAGNYY1872 pKa = 9.62DD1873 pKa = 3.63YY1874 pKa = 11.24QLNATGLAKK1883 pKa = 10.26VQSVVGPNYY1892 pKa = 9.58TVVLGGSGTVSISQVVATTVLSDD1915 pKa = 3.75GGFTFDD1921 pKa = 3.52GTTKK1925 pKa = 10.64ASEE1928 pKa = 4.37VTNLTASVVGQTVEE1942 pKa = 4.67LDD1944 pKa = 3.31SDD1946 pKa = 4.65DD1947 pKa = 5.94IIVTDD1952 pKa = 5.1DD1953 pKa = 3.28GSAAQDD1959 pKa = 2.63SWARR1963 pKa = 3.58
MM1 pKa = 7.95LSPQVQNVAASTATAGSTTEE21 pKa = 4.04EE22 pKa = 4.05TSGITNTSVAHH33 pKa = 6.36SSADD37 pKa = 3.34VKK39 pKa = 11.3ASVASTSSATVVNTGNDD56 pKa = 3.17SSTATSATSTASASTSVSAVASATNGSAEE85 pKa = 4.17SLAATSAATAATTSVAVDD103 pKa = 3.94DD104 pKa = 4.93SSAALPASSAATAGAEE120 pKa = 4.29TSSATSGSSAASAQTVTSVAHH141 pKa = 4.67QALRR145 pKa = 11.84TRR147 pKa = 11.84SLASSAAPADD157 pKa = 4.05SEE159 pKa = 4.48VTTAINSGAANAPATVSYY177 pKa = 10.87HH178 pKa = 6.11NGSTQPTISVMVSGNFQAGDD198 pKa = 3.66VVTVYY203 pKa = 11.26LDD205 pKa = 3.49TTAMSVTAATTVKK218 pKa = 10.4GATEE222 pKa = 4.3SNVDD226 pKa = 2.93VSTNDD231 pKa = 2.63GRR233 pKa = 11.84TVKK236 pKa = 10.26GAQYY240 pKa = 10.61QFSSDD245 pKa = 3.57YY246 pKa = 10.94NGAFPVVFVLNGTADD261 pKa = 3.65DD262 pKa = 4.34QINNGTSTATIAVDD276 pKa = 3.22VDD278 pKa = 3.77KK279 pKa = 11.5NGTIVATNSVVLDD292 pKa = 3.58KK293 pKa = 10.82SYY295 pKa = 10.48TMTQPNDD302 pKa = 3.62TIRR305 pKa = 11.84RR306 pKa = 11.84NILNGTNGVTAGVIAQGAVTSEE328 pKa = 3.92LTLRR332 pKa = 11.84AQLQGLQYY340 pKa = 10.69FGDD343 pKa = 3.94YY344 pKa = 8.83STLVNSSPYY353 pKa = 10.35EE354 pKa = 4.0KK355 pKa = 9.79LTAASTVTFDD365 pKa = 6.0FSDD368 pKa = 3.51QLKK371 pKa = 10.21VDD373 pKa = 4.02SAVLSDD379 pKa = 4.17DD380 pKa = 3.84AVATGVTLTRR390 pKa = 11.84SGNSVTLTLPSGLTLAAISKK410 pKa = 9.8LAGYY414 pKa = 8.69LQVGLLPVTVPTTSTRR430 pKa = 11.84YY431 pKa = 9.63VLPTLTATWTIDD443 pKa = 3.13GQTNTVTEE451 pKa = 4.51PGNSGSSHH459 pKa = 6.3ANEE462 pKa = 3.7FAYY465 pKa = 10.64NIQTYY470 pKa = 7.62TAVGTVTVSSSSSYY484 pKa = 11.08LGLLQQGTDD493 pKa = 3.19NDD495 pKa = 4.39YY496 pKa = 10.83GLQWVVANEE505 pKa = 4.32SNFASGTTTTGSLSITSSPDD525 pKa = 2.86NSVGITSVNGLDD537 pKa = 3.88GNYY540 pKa = 7.86LTSAGLSKK548 pKa = 10.96NLTITFTTADD558 pKa = 3.63GQSQSFEE565 pKa = 4.31YY566 pKa = 11.11VDD568 pKa = 3.6GTTSYY573 pKa = 11.08VATVGSATDD582 pKa = 3.68PVTQVQIVDD591 pKa = 4.04HH592 pKa = 6.77AAQTAPYY599 pKa = 7.97VTGSISHH606 pKa = 6.1LQVGLIAYY614 pKa = 9.29GDD616 pKa = 3.77EE617 pKa = 4.31KK618 pKa = 9.81TTVTATADD626 pKa = 3.17TSLYY630 pKa = 11.0DD631 pKa = 3.67GDD633 pKa = 4.31GKK635 pKa = 11.11LVLDD639 pKa = 4.43TASSTDD645 pKa = 3.02SDD647 pKa = 4.14VVRR650 pKa = 11.84EE651 pKa = 4.01RR652 pKa = 11.84TVTKK656 pKa = 9.67TYY658 pKa = 10.42YY659 pKa = 8.19PQPSIVLSAGSNGVDD674 pKa = 3.01NQQGNTAQQIYY685 pKa = 10.71NPGDD689 pKa = 3.55EE690 pKa = 4.4LTEE693 pKa = 4.31SVNLLDD699 pKa = 4.95GGTNSGSASDD709 pKa = 3.52QVGRR713 pKa = 11.84IGNGDD718 pKa = 3.51GKK720 pKa = 10.56QPAVEE725 pKa = 4.67AVAAQNPMTIVVVAPTNTDD744 pKa = 3.49FADD747 pKa = 4.09DD748 pKa = 3.76ATLLSTLNKK757 pKa = 10.16SDD759 pKa = 3.33ISSSGDD765 pKa = 3.41LEE767 pKa = 4.2ITHH770 pKa = 7.15LDD772 pKa = 3.82DD773 pKa = 3.65VDD775 pKa = 3.81GRR777 pKa = 11.84PVIAITGVDD786 pKa = 3.67HH787 pKa = 5.71YY788 pKa = 9.22TASSRR793 pKa = 11.84LPIVLQVNADD803 pKa = 4.0AIGGSTISALNTIYY817 pKa = 11.2VMPSAVLSQYY827 pKa = 11.71NNGTILTSDD836 pKa = 3.59GSLATDD842 pKa = 3.72WATGNMYY849 pKa = 10.68NLSTGGSQLGTIITASSFTANASITGSADD878 pKa = 2.58SSAQVQTGTINVDD891 pKa = 3.03DD892 pKa = 4.83DD893 pKa = 3.67QATGKK898 pKa = 6.27TTLYY902 pKa = 10.86NGTTNPVTDD911 pKa = 3.41TLTTVNLVDD920 pKa = 4.47ADD922 pKa = 3.81GNQYY926 pKa = 8.16ITLTGPVQVVDD937 pKa = 3.9DD938 pKa = 4.8AGNTVAAAVTYY949 pKa = 7.38YY950 pKa = 11.03QNGAVVTDD958 pKa = 3.93PTQADD963 pKa = 3.97SFTVSGVSLAKK974 pKa = 9.97KK975 pKa = 10.1GSYY978 pKa = 10.44AVISYY983 pKa = 10.24GLTVVPSQLAGALTNKK999 pKa = 7.7TWAYY1003 pKa = 6.81QTSTQGYY1010 pKa = 5.64TAGTVATGNLVPLGAAISNTLVLQAIRR1037 pKa = 11.84DD1038 pKa = 4.02LTDD1041 pKa = 2.43TWYY1044 pKa = 10.33VAQRR1048 pKa = 11.84KK1049 pKa = 9.29ADD1051 pKa = 3.46GTYY1054 pKa = 10.46VVDD1057 pKa = 3.76SAVAAVTTTGVVGRR1071 pKa = 11.84IYY1073 pKa = 10.86NGYY1076 pKa = 7.5TLMGTSAADD1085 pKa = 3.1VGQAYY1090 pKa = 10.28GDD1092 pKa = 4.15DD1093 pKa = 3.88GAVLVGVYY1101 pKa = 10.29DD1102 pKa = 3.6ATTGQMVADD1111 pKa = 4.65GSMITYY1117 pKa = 10.32AEE1119 pKa = 4.11QVNSDD1124 pKa = 3.57GTITGHH1130 pKa = 5.76TYY1132 pKa = 11.26YY1133 pKa = 10.95LVTEE1137 pKa = 4.12QDD1139 pKa = 3.42VATKK1143 pKa = 10.79LNDD1146 pKa = 3.13VTVTYY1151 pKa = 10.55NGQTAASEE1159 pKa = 4.47VAGLQATANKK1169 pKa = 9.77VNVGLTSDD1177 pKa = 5.21DD1178 pKa = 3.21ITVVKK1183 pKa = 10.7DD1184 pKa = 3.74GINAGTYY1191 pKa = 8.91TYY1193 pKa = 11.03QLNAQGLAKK1202 pKa = 10.37LQAAVGNDD1210 pKa = 3.83AIVTDD1215 pKa = 3.92VGVTGTITIQPAVTTVTLNPGALTYY1240 pKa = 10.62DD1241 pKa = 3.14GRR1243 pKa = 11.84TPASGASLTATVAGQTLKK1261 pKa = 11.06LNSGDD1266 pKa = 3.44VTVADD1271 pKa = 4.78DD1272 pKa = 5.08GIDD1275 pKa = 3.35VGTYY1279 pKa = 9.01TYY1281 pKa = 11.18QLTAAGLSSLQSQVGSNYY1299 pKa = 9.79VVTTGTPGMIVISPEE1314 pKa = 3.83QMPVTLSSGSLVYY1327 pKa = 10.42DD1328 pKa = 4.38GKK1330 pKa = 9.4TKK1332 pKa = 10.87ASGAAVIATVNGEE1345 pKa = 4.31PLTLNDD1351 pKa = 5.4DD1352 pKa = 5.25DD1353 pKa = 4.2ITVTNDD1359 pKa = 3.87GINVGTYY1366 pKa = 10.1NYY1368 pKa = 10.43QLNQTGLEE1376 pKa = 4.03KK1377 pKa = 10.83LQTLVGDD1384 pKa = 4.13DD1385 pKa = 3.66FTVVVGDD1392 pKa = 4.47PGTITITPAVVDD1404 pKa = 3.71VVLNDD1409 pKa = 3.82GSFTYY1414 pKa = 10.46DD1415 pKa = 2.78GNTAASAAKK1424 pKa = 10.42GLVATVNGTPVDD1436 pKa = 4.38LDD1438 pKa = 3.54NSDD1441 pKa = 4.72IEE1443 pKa = 4.47VTDD1446 pKa = 4.59DD1447 pKa = 4.67GVDD1450 pKa = 3.22AGNYY1454 pKa = 9.6AYY1456 pKa = 10.49QLNDD1460 pKa = 3.17AGLAKK1465 pKa = 10.29VQLVVGSNYY1474 pKa = 9.37QVSVAVTGTITITPASITAALSDD1497 pKa = 4.11GGFTYY1502 pKa = 10.69DD1503 pKa = 3.36GATKK1507 pKa = 10.35ASEE1510 pKa = 4.28ATGLTATANSQAVDD1524 pKa = 3.71LSGEE1528 pKa = 4.42DD1529 pKa = 3.61IEE1531 pKa = 4.63VTNDD1535 pKa = 2.79GTGAGNYY1542 pKa = 9.65AYY1544 pKa = 10.13QLSDD1548 pKa = 3.09AGLAKK1553 pKa = 10.06VQSVVGTNYY1562 pKa = 9.75TVVLGGSGTVTIAQVVATTVXQSAVGPNYY1591 pKa = 9.78TVGVGDD1597 pKa = 4.13TGTITITPASITTTLSDD1614 pKa = 4.17GGFTYY1619 pKa = 10.71DD1620 pKa = 3.48GTTKK1624 pKa = 10.93ASGAPKK1630 pKa = 9.24LTATVVGQTVEE1641 pKa = 4.48LDD1643 pKa = 3.52SSDD1646 pKa = 4.41VDD1648 pKa = 3.8VTDD1651 pKa = 3.89NGSAAGDD1658 pKa = 3.44YY1659 pKa = 10.62VYY1661 pKa = 10.92QLNANGLEE1669 pKa = 4.38KK1670 pKa = 10.81VQSAAGANYY1679 pKa = 9.17IVRR1682 pKa = 11.84VGDD1685 pKa = 3.34TGTITIAKK1693 pKa = 9.71APATTTLSDD1702 pKa = 3.74GGFTYY1707 pKa = 10.71DD1708 pKa = 3.48GTTKK1712 pKa = 10.93ASGAPKK1718 pKa = 9.24LTATVVGQTVEE1729 pKa = 4.67LDD1731 pKa = 3.4SSDD1734 pKa = 3.09IDD1736 pKa = 4.18VINDD1740 pKa = 4.49AIAVGSYY1747 pKa = 10.35DD1748 pKa = 3.6YY1749 pKa = 11.14QLNATGLAKK1758 pKa = 10.47VQSAAGGNYY1767 pKa = 8.62TVGFGDD1773 pKa = 4.4AGTITITPASITTTLSDD1790 pKa = 4.17GGFTYY1795 pKa = 10.71DD1796 pKa = 3.48GTTKK1800 pKa = 10.93ASGAPKK1806 pKa = 9.34XFGDD1810 pKa = 4.38AGTITITPASITTTLSDD1827 pKa = 4.17GGFTYY1832 pKa = 10.71DD1833 pKa = 3.48GTTKK1837 pKa = 10.93ASGAPKK1843 pKa = 9.25LTATVIGQTVDD1854 pKa = 4.05LDD1856 pKa = 3.86SSDD1859 pKa = 4.12IDD1861 pKa = 3.52ITNDD1865 pKa = 2.45ATTAGNYY1872 pKa = 9.62DD1873 pKa = 3.63YY1874 pKa = 11.24QLNATGLAKK1883 pKa = 10.26VQSVVGPNYY1892 pKa = 9.58TVVLGGSGTVSISQVVATTVLSDD1915 pKa = 3.75GGFTFDD1921 pKa = 3.52GTTKK1925 pKa = 10.64ASEE1928 pKa = 4.37VTNLTASVVGQTVEE1942 pKa = 4.67LDD1944 pKa = 3.31SDD1946 pKa = 4.65DD1947 pKa = 5.94IIVTDD1952 pKa = 5.1DD1953 pKa = 3.28GSAAQDD1959 pKa = 2.63SWARR1963 pKa = 3.58
Molecular weight: 197.71 kDa
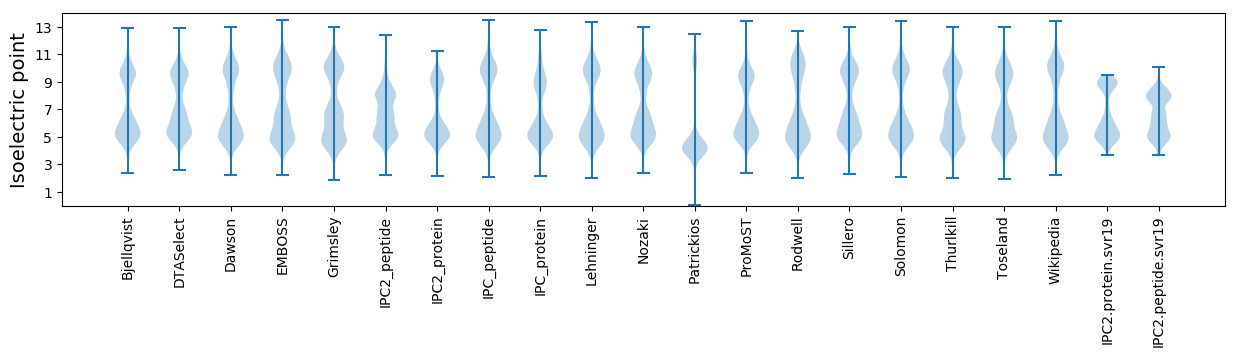
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R2CGU8|A0A0R2CGU8_9LACO 50S ribosomal protein L30 OS=Lactobacillus thailandensis DSM 22698 = JCM 13996 OX=1423810 GN=rpmD PE=3 SV=1
MM1 pKa = 5.72TTKK4 pKa = 9.85RR5 pKa = 11.84TFQPKK10 pKa = 8.26KK11 pKa = 7.4RR12 pKa = 11.84HH13 pKa = 5.36RR14 pKa = 11.84ARR16 pKa = 11.84VHH18 pKa = 6.05GFMKK22 pKa = 10.54RR23 pKa = 11.84MSTKK27 pKa = 10.23NGRR30 pKa = 11.84HH31 pKa = 3.67VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.16GRR41 pKa = 11.84KK42 pKa = 9.04KK43 pKa = 9.67LTAA46 pKa = 4.2
MM1 pKa = 5.72TTKK4 pKa = 9.85RR5 pKa = 11.84TFQPKK10 pKa = 8.26KK11 pKa = 7.4RR12 pKa = 11.84HH13 pKa = 5.36RR14 pKa = 11.84ARR16 pKa = 11.84VHH18 pKa = 6.05GFMKK22 pKa = 10.54RR23 pKa = 11.84MSTKK27 pKa = 10.23NGRR30 pKa = 11.84HH31 pKa = 3.67VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.16GRR41 pKa = 11.84KK42 pKa = 9.04KK43 pKa = 9.67LTAA46 pKa = 4.2
Molecular weight: 5.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
581517 |
46 |
2509 |
312.6 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.422 ± 0.079 | 0.416 ± 0.012 |
6.111 ± 0.06 | 4.387 ± 0.053 |
3.794 ± 0.037 | 7.368 ± 0.053 |
2.598 ± 0.031 | 5.897 ± 0.048 |
4.305 ± 0.062 | 9.561 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.749 ± 0.03 | 3.965 ± 0.04 |
3.927 ± 0.038 | 4.673 ± 0.042 |
5.063 ± 0.047 | 5.334 ± 0.077 |
6.912 ± 0.078 | 8.147 ± 0.047 |
1.104 ± 0.022 | 3.264 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
