
Simian retrovirus Y
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; unclassified Retroviridae
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
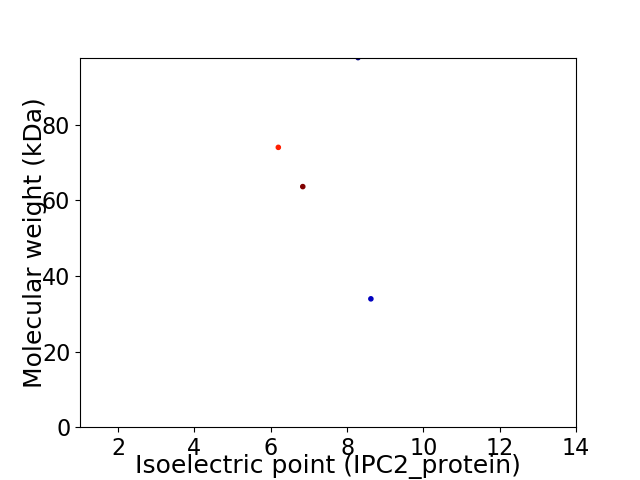
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0MZL7|L0MZL7_9RETR Envelope protein OS=Simian retrovirus Y OX=980635 GN=env PE=4 SV=1
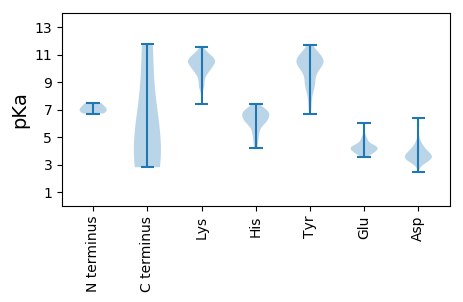
MM1 pKa = 6.93GQEE4 pKa = 4.61LSQHH8 pKa = 6.05DD9 pKa = 5.05LYY11 pKa = 11.71VDD13 pKa = 3.59QLKK16 pKa = 10.53KK17 pKa = 10.36ALKK20 pKa = 10.08ARR22 pKa = 11.84GVKK25 pKa = 9.66VKK27 pKa = 10.77YY28 pKa = 10.54ADD30 pKa = 3.61LLKK33 pKa = 11.07FFDD36 pKa = 4.81FVKK39 pKa = 9.86DD40 pKa = 3.41TCPWFPQEE48 pKa = 3.67GTIDD52 pKa = 3.17IKK54 pKa = 10.13RR55 pKa = 11.84WRR57 pKa = 11.84RR58 pKa = 11.84VGDD61 pKa = 3.88CFQDD65 pKa = 3.72YY66 pKa = 10.54YY67 pKa = 10.97KK68 pKa = 10.65TFGPEE73 pKa = 3.94KK74 pKa = 10.38VPVTAFSYY82 pKa = 10.07WNLIKK87 pKa = 10.52EE88 pKa = 4.72LIDD91 pKa = 3.58KK92 pKa = 11.04KK93 pKa = 10.57EE94 pKa = 3.93DD95 pKa = 3.45HH96 pKa = 6.89PQVLAAVTQTEE107 pKa = 4.32EE108 pKa = 4.02LLKK111 pKa = 10.31TGSQSDD117 pKa = 4.2LQHH120 pKa = 6.76LPPDD124 pKa = 4.34PEE126 pKa = 4.22TDD128 pKa = 4.57LISLDD133 pKa = 3.76SDD135 pKa = 3.72EE136 pKa = 5.37EE137 pKa = 4.4GAKK140 pKa = 10.52APSIKK145 pKa = 10.23DD146 pKa = 3.32CKK148 pKa = 8.53TVRR151 pKa = 11.84NKK153 pKa = 10.14KK154 pKa = 9.18PKK156 pKa = 9.96RR157 pKa = 11.84FPVLLTSQEE166 pKa = 4.27SKK168 pKa = 11.28NDD170 pKa = 3.74DD171 pKa = 4.15NDD173 pKa = 4.48PDD175 pKa = 5.92DD176 pKa = 6.28DD177 pKa = 5.91DD178 pKa = 5.84PDD180 pKa = 4.0PQEE183 pKa = 4.81VNWDD187 pKa = 3.9DD188 pKa = 4.26LADD191 pKa = 3.74AAARR195 pKa = 11.84YY196 pKa = 8.02HH197 pKa = 6.25NPDD200 pKa = 2.99WPPFLTRR207 pKa = 11.84PPPYY211 pKa = 10.52NKK213 pKa = 9.84AAPSAPEE220 pKa = 3.67VMAVVNPKK228 pKa = 10.56EE229 pKa = 3.84EE230 pKa = 4.31LKK232 pKa = 11.05EE233 pKa = 4.15KK234 pKa = 10.21IAQLEE239 pKa = 4.17EE240 pKa = 4.15QIKK243 pKa = 10.86LEE245 pKa = 4.18EE246 pKa = 4.12LHH248 pKa = 6.4QSLISRR254 pKa = 11.84LQKK257 pKa = 10.7LKK259 pKa = 10.02TGNEE263 pKa = 3.69RR264 pKa = 11.84VTNLDD269 pKa = 3.24NAGGFPRR276 pKa = 11.84TPHH279 pKa = 6.86WPRR282 pKa = 11.84QHH284 pKa = 5.53VPKK287 pKa = 10.5GKK289 pKa = 10.24SYY291 pKa = 10.85TNIEE295 pKa = 4.33RR296 pKa = 11.84EE297 pKa = 4.2PSPKK301 pKa = 10.32DD302 pKa = 2.95IFPVTEE308 pKa = 3.98TVDD311 pKa = 3.43AQGQAWRR318 pKa = 11.84HH319 pKa = 5.12HH320 pKa = 6.22NGFDD324 pKa = 3.27FTVIKK329 pKa = 9.85EE330 pKa = 4.31LKK332 pKa = 8.64TAASQYY338 pKa = 10.77GATAPYY344 pKa = 8.96TLAIVEE350 pKa = 4.48SVADD354 pKa = 3.63NWLTPTDD361 pKa = 3.42WNTLVRR367 pKa = 11.84AVLSGGDD374 pKa = 3.36HH375 pKa = 6.8LLWKK379 pKa = 10.66SEE381 pKa = 3.58FFEE384 pKa = 4.44NCRR387 pKa = 11.84EE388 pKa = 3.98TAKK391 pKa = 10.61RR392 pKa = 11.84NQQAGNGWDD401 pKa = 3.64FDD403 pKa = 4.06MLTGSGNYY411 pKa = 10.15SNTDD415 pKa = 2.46AQMQYY420 pKa = 11.44DD421 pKa = 4.17PGLFAQIQAAAIKK434 pKa = 9.62AWRR437 pKa = 11.84KK438 pKa = 9.78LPVKK442 pKa = 9.92GDD444 pKa = 3.67PGASLTGVKK453 pKa = 9.76QGPDD457 pKa = 3.08EE458 pKa = 4.48PFSEE462 pKa = 4.91FVHH465 pKa = 7.31RR466 pKa = 11.84LITTAGRR473 pKa = 11.84IFGNAEE479 pKa = 3.61AGVDD483 pKa = 3.84YY484 pKa = 10.87VKK486 pKa = 10.86QLAYY490 pKa = 10.49EE491 pKa = 4.22NANPACQAAIRR502 pKa = 11.84PYY504 pKa = 10.38RR505 pKa = 11.84KK506 pKa = 8.54KK507 pKa = 9.28TDD509 pKa = 3.14LTGYY513 pKa = 9.9IRR515 pKa = 11.84LCSDD519 pKa = 3.8IGPSYY524 pKa = 10.24QQGLAMAAAFSGQTVKK540 pKa = 11.11DD541 pKa = 3.64FLNNKK546 pKa = 8.72HH547 pKa = 5.96RR548 pKa = 11.84EE549 pKa = 3.78KK550 pKa = 10.93GGCFKK555 pKa = 10.7CGKK558 pKa = 9.61AGHH561 pKa = 6.02FAKK564 pKa = 10.59NCQEE568 pKa = 4.14NIKK571 pKa = 10.95NPGTKK576 pKa = 9.98VPGVCPRR583 pKa = 11.84CKK585 pKa = 9.94RR586 pKa = 11.84GKK588 pKa = 9.48HH589 pKa = 4.71WANEE593 pKa = 4.47CKK595 pKa = 10.49SKK597 pKa = 9.96TDD599 pKa = 3.38SQGNPLPPHH608 pKa = 6.29QGNGWRR614 pKa = 11.84GQPQAPKK621 pKa = 9.09QAYY624 pKa = 8.28GAVSFVPANNNNPFQNLLEE643 pKa = 4.1PHH645 pKa = 6.15QEE647 pKa = 4.15VQDD650 pKa = 3.85WTSVPPPTQYY660 pKa = 11.76
MM1 pKa = 6.93GQEE4 pKa = 4.61LSQHH8 pKa = 6.05DD9 pKa = 5.05LYY11 pKa = 11.71VDD13 pKa = 3.59QLKK16 pKa = 10.53KK17 pKa = 10.36ALKK20 pKa = 10.08ARR22 pKa = 11.84GVKK25 pKa = 9.66VKK27 pKa = 10.77YY28 pKa = 10.54ADD30 pKa = 3.61LLKK33 pKa = 11.07FFDD36 pKa = 4.81FVKK39 pKa = 9.86DD40 pKa = 3.41TCPWFPQEE48 pKa = 3.67GTIDD52 pKa = 3.17IKK54 pKa = 10.13RR55 pKa = 11.84WRR57 pKa = 11.84RR58 pKa = 11.84VGDD61 pKa = 3.88CFQDD65 pKa = 3.72YY66 pKa = 10.54YY67 pKa = 10.97KK68 pKa = 10.65TFGPEE73 pKa = 3.94KK74 pKa = 10.38VPVTAFSYY82 pKa = 10.07WNLIKK87 pKa = 10.52EE88 pKa = 4.72LIDD91 pKa = 3.58KK92 pKa = 11.04KK93 pKa = 10.57EE94 pKa = 3.93DD95 pKa = 3.45HH96 pKa = 6.89PQVLAAVTQTEE107 pKa = 4.32EE108 pKa = 4.02LLKK111 pKa = 10.31TGSQSDD117 pKa = 4.2LQHH120 pKa = 6.76LPPDD124 pKa = 4.34PEE126 pKa = 4.22TDD128 pKa = 4.57LISLDD133 pKa = 3.76SDD135 pKa = 3.72EE136 pKa = 5.37EE137 pKa = 4.4GAKK140 pKa = 10.52APSIKK145 pKa = 10.23DD146 pKa = 3.32CKK148 pKa = 8.53TVRR151 pKa = 11.84NKK153 pKa = 10.14KK154 pKa = 9.18PKK156 pKa = 9.96RR157 pKa = 11.84FPVLLTSQEE166 pKa = 4.27SKK168 pKa = 11.28NDD170 pKa = 3.74DD171 pKa = 4.15NDD173 pKa = 4.48PDD175 pKa = 5.92DD176 pKa = 6.28DD177 pKa = 5.91DD178 pKa = 5.84PDD180 pKa = 4.0PQEE183 pKa = 4.81VNWDD187 pKa = 3.9DD188 pKa = 4.26LADD191 pKa = 3.74AAARR195 pKa = 11.84YY196 pKa = 8.02HH197 pKa = 6.25NPDD200 pKa = 2.99WPPFLTRR207 pKa = 11.84PPPYY211 pKa = 10.52NKK213 pKa = 9.84AAPSAPEE220 pKa = 3.67VMAVVNPKK228 pKa = 10.56EE229 pKa = 3.84EE230 pKa = 4.31LKK232 pKa = 11.05EE233 pKa = 4.15KK234 pKa = 10.21IAQLEE239 pKa = 4.17EE240 pKa = 4.15QIKK243 pKa = 10.86LEE245 pKa = 4.18EE246 pKa = 4.12LHH248 pKa = 6.4QSLISRR254 pKa = 11.84LQKK257 pKa = 10.7LKK259 pKa = 10.02TGNEE263 pKa = 3.69RR264 pKa = 11.84VTNLDD269 pKa = 3.24NAGGFPRR276 pKa = 11.84TPHH279 pKa = 6.86WPRR282 pKa = 11.84QHH284 pKa = 5.53VPKK287 pKa = 10.5GKK289 pKa = 10.24SYY291 pKa = 10.85TNIEE295 pKa = 4.33RR296 pKa = 11.84EE297 pKa = 4.2PSPKK301 pKa = 10.32DD302 pKa = 2.95IFPVTEE308 pKa = 3.98TVDD311 pKa = 3.43AQGQAWRR318 pKa = 11.84HH319 pKa = 5.12HH320 pKa = 6.22NGFDD324 pKa = 3.27FTVIKK329 pKa = 9.85EE330 pKa = 4.31LKK332 pKa = 8.64TAASQYY338 pKa = 10.77GATAPYY344 pKa = 8.96TLAIVEE350 pKa = 4.48SVADD354 pKa = 3.63NWLTPTDD361 pKa = 3.42WNTLVRR367 pKa = 11.84AVLSGGDD374 pKa = 3.36HH375 pKa = 6.8LLWKK379 pKa = 10.66SEE381 pKa = 3.58FFEE384 pKa = 4.44NCRR387 pKa = 11.84EE388 pKa = 3.98TAKK391 pKa = 10.61RR392 pKa = 11.84NQQAGNGWDD401 pKa = 3.64FDD403 pKa = 4.06MLTGSGNYY411 pKa = 10.15SNTDD415 pKa = 2.46AQMQYY420 pKa = 11.44DD421 pKa = 4.17PGLFAQIQAAAIKK434 pKa = 9.62AWRR437 pKa = 11.84KK438 pKa = 9.78LPVKK442 pKa = 9.92GDD444 pKa = 3.67PGASLTGVKK453 pKa = 9.76QGPDD457 pKa = 3.08EE458 pKa = 4.48PFSEE462 pKa = 4.91FVHH465 pKa = 7.31RR466 pKa = 11.84LITTAGRR473 pKa = 11.84IFGNAEE479 pKa = 3.61AGVDD483 pKa = 3.84YY484 pKa = 10.87VKK486 pKa = 10.86QLAYY490 pKa = 10.49EE491 pKa = 4.22NANPACQAAIRR502 pKa = 11.84PYY504 pKa = 10.38RR505 pKa = 11.84KK506 pKa = 8.54KK507 pKa = 9.28TDD509 pKa = 3.14LTGYY513 pKa = 9.9IRR515 pKa = 11.84LCSDD519 pKa = 3.8IGPSYY524 pKa = 10.24QQGLAMAAAFSGQTVKK540 pKa = 11.11DD541 pKa = 3.64FLNNKK546 pKa = 8.72HH547 pKa = 5.96RR548 pKa = 11.84EE549 pKa = 3.78KK550 pKa = 10.93GGCFKK555 pKa = 10.7CGKK558 pKa = 9.61AGHH561 pKa = 6.02FAKK564 pKa = 10.59NCQEE568 pKa = 4.14NIKK571 pKa = 10.95NPGTKK576 pKa = 9.98VPGVCPRR583 pKa = 11.84CKK585 pKa = 9.94RR586 pKa = 11.84GKK588 pKa = 9.48HH589 pKa = 4.71WANEE593 pKa = 4.47CKK595 pKa = 10.49SKK597 pKa = 9.96TDD599 pKa = 3.38SQGNPLPPHH608 pKa = 6.29QGNGWRR614 pKa = 11.84GQPQAPKK621 pKa = 9.09QAYY624 pKa = 8.28GAVSFVPANNNNPFQNLLEE643 pKa = 4.1PHH645 pKa = 6.15QEE647 pKa = 4.15VQDD650 pKa = 3.85WTSVPPPTQYY660 pKa = 11.76
Molecular weight: 73.96 kDa
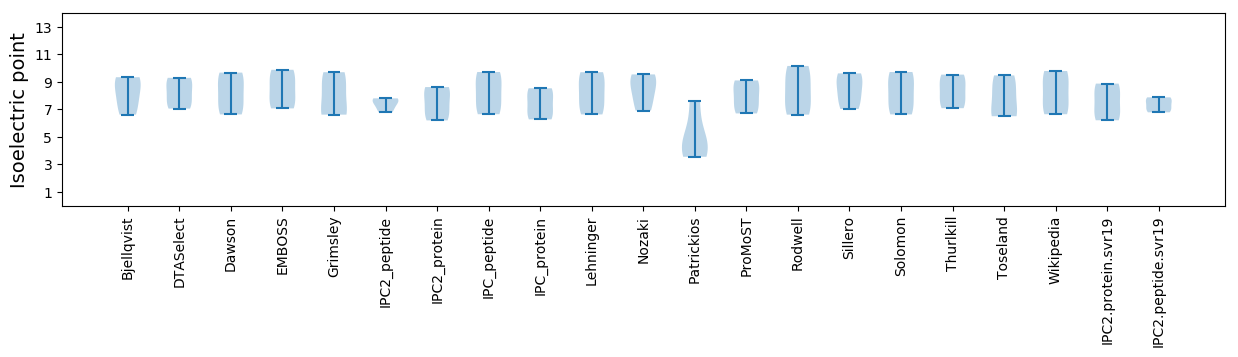
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0MXX7|L0MXX7_9RETR Gag protein OS=Simian retrovirus Y OX=980635 GN=gag PE=4 SV=1
SS1 pKa = 7.13RR2 pKa = 11.84KK3 pKa = 8.55STPTPSGKK11 pKa = 9.56RR12 pKa = 11.84VEE14 pKa = 4.81GPAPGPEE21 pKa = 3.72TSLWGGQLCPSQQQQSISEE40 pKa = 4.33LARR43 pKa = 11.84ATPGSAGLDD52 pKa = 3.4LSSASHH58 pKa = 5.85TVLTPEE64 pKa = 5.01MGPQALGTGIYY75 pKa = 10.61GPLPTNTFGLIVGRR89 pKa = 11.84SSITMKK95 pKa = 10.56GLQVFPGIIDD105 pKa = 3.44NDD107 pKa = 3.65YY108 pKa = 9.78TGEE111 pKa = 3.96IKK113 pKa = 10.46IMAKK117 pKa = 10.17ALNNIITIPQGDD129 pKa = 4.5RR130 pKa = 11.84IAQLVLLPLIKK141 pKa = 9.87TSNKK145 pKa = 8.09VQQSFRR151 pKa = 11.84GQKK154 pKa = 10.43GFGSSDD160 pKa = 3.44IYY162 pKa = 10.15WVQPITNKK170 pKa = 10.15KK171 pKa = 9.74PSLTLWLDD179 pKa = 3.53GKK181 pKa = 11.13SFTGLMDD188 pKa = 3.6TGADD192 pKa = 3.3VTIIKK197 pKa = 10.34QEE199 pKa = 3.87DD200 pKa = 4.07WPPNWPTTEE209 pKa = 3.83TLTNLRR215 pKa = 11.84GIGQSNNPRR224 pKa = 11.84QSSKK228 pKa = 11.03YY229 pKa = 8.55LTWKK233 pKa = 10.59DD234 pKa = 3.32KK235 pKa = 11.17EE236 pKa = 4.36NNTGLIKK243 pKa = 10.5PFIIPNLPVNLWGRR257 pKa = 11.84DD258 pKa = 3.49LLSQMKK264 pKa = 10.38IMMCSPNDD272 pKa = 3.42VVTAQMLAQGYY283 pKa = 9.59SPGKK287 pKa = 10.3GLGKK291 pKa = 10.29NEE293 pKa = 4.59DD294 pKa = 4.75GILQPIPNLGQFDD307 pKa = 4.08KK308 pKa = 11.24KK309 pKa = 11.24GLGNFF314 pKa = 3.79
SS1 pKa = 7.13RR2 pKa = 11.84KK3 pKa = 8.55STPTPSGKK11 pKa = 9.56RR12 pKa = 11.84VEE14 pKa = 4.81GPAPGPEE21 pKa = 3.72TSLWGGQLCPSQQQQSISEE40 pKa = 4.33LARR43 pKa = 11.84ATPGSAGLDD52 pKa = 3.4LSSASHH58 pKa = 5.85TVLTPEE64 pKa = 5.01MGPQALGTGIYY75 pKa = 10.61GPLPTNTFGLIVGRR89 pKa = 11.84SSITMKK95 pKa = 10.56GLQVFPGIIDD105 pKa = 3.44NDD107 pKa = 3.65YY108 pKa = 9.78TGEE111 pKa = 3.96IKK113 pKa = 10.46IMAKK117 pKa = 10.17ALNNIITIPQGDD129 pKa = 4.5RR130 pKa = 11.84IAQLVLLPLIKK141 pKa = 9.87TSNKK145 pKa = 8.09VQQSFRR151 pKa = 11.84GQKK154 pKa = 10.43GFGSSDD160 pKa = 3.44IYY162 pKa = 10.15WVQPITNKK170 pKa = 10.15KK171 pKa = 9.74PSLTLWLDD179 pKa = 3.53GKK181 pKa = 11.13SFTGLMDD188 pKa = 3.6TGADD192 pKa = 3.3VTIIKK197 pKa = 10.34QEE199 pKa = 3.87DD200 pKa = 4.07WPPNWPTTEE209 pKa = 3.83TLTNLRR215 pKa = 11.84GIGQSNNPRR224 pKa = 11.84QSSKK228 pKa = 11.03YY229 pKa = 8.55LTWKK233 pKa = 10.59DD234 pKa = 3.32KK235 pKa = 11.17EE236 pKa = 4.36NNTGLIKK243 pKa = 10.5PFIIPNLPVNLWGRR257 pKa = 11.84DD258 pKa = 3.49LLSQMKK264 pKa = 10.38IMMCSPNDD272 pKa = 3.42VVTAQMLAQGYY283 pKa = 9.59SPGKK287 pKa = 10.3GLGKK291 pKa = 10.29NEE293 pKa = 4.59DD294 pKa = 4.75GILQPIPNLGQFDD307 pKa = 4.08KK308 pKa = 11.24KK309 pKa = 11.24GLGNFF314 pKa = 3.79
Molecular weight: 33.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2423 |
314 |
864 |
605.8 |
67.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.181 ± 0.845 | 2.064 ± 0.544 |
5.489 ± 0.561 | 3.673 ± 0.665 |
3.673 ± 0.387 | 6.48 ± 1.178 |
2.435 ± 0.542 | 6.191 ± 0.99 |
6.521 ± 0.759 | 10.194 ± 0.99 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.197 ± 0.345 | 5.076 ± 0.221 |
7.222 ± 0.475 | 6.356 ± 0.257 |
3.591 ± 0.222 | 6.067 ± 0.939 |
6.686 ± 0.464 | 5.241 ± 0.501 |
1.94 ± 0.275 | 2.724 ± 0.274 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
