
Carrot Ch virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Chordovirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
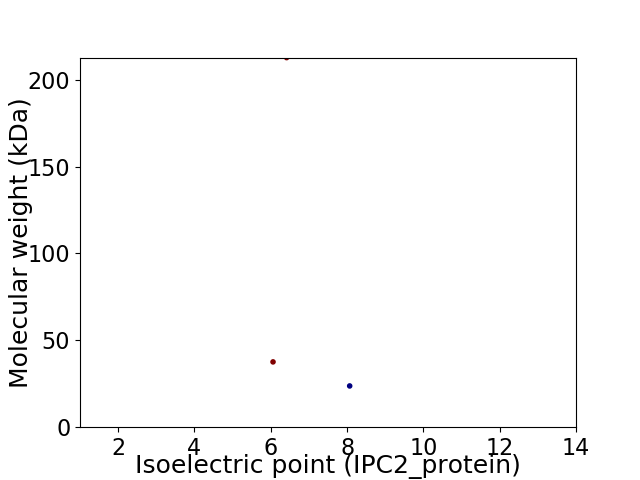
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0P710|A0A0A0P710_9VIRU ORF1 OS=Carrot Ch virus 1 OX=1425363 PE=4 SV=1
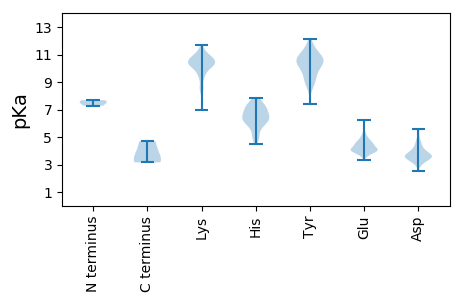
MM1 pKa = 7.52SLVDD5 pKa = 3.78VNKK8 pKa = 9.99FCSLVSGGKK17 pKa = 9.85SSTEE21 pKa = 4.32AISSSNIYY29 pKa = 10.58GDD31 pKa = 4.31SSWLKK36 pKa = 10.29LKK38 pKa = 10.87NITAIKK44 pKa = 10.2KK45 pKa = 9.69FEE47 pKa = 4.36TNIEE51 pKa = 4.12LEE53 pKa = 4.43LEE55 pKa = 4.77DD56 pKa = 5.11GDD58 pKa = 4.4SKK60 pKa = 11.46LVIPNLPLISDD71 pKa = 3.81SEE73 pKa = 3.9IRR75 pKa = 11.84AVKK78 pKa = 10.61KK79 pKa = 10.63NMPSVNFVHH88 pKa = 6.96LGGIVVSIQALFAANKK104 pKa = 8.49GVKK107 pKa = 8.37GTAVLVDD114 pKa = 3.68RR115 pKa = 11.84RR116 pKa = 11.84WRR118 pKa = 11.84NLDD121 pKa = 3.09QAIIGSFAFNLDD133 pKa = 2.89KK134 pKa = 11.02RR135 pKa = 11.84RR136 pKa = 11.84ADD138 pKa = 3.54FMMRR142 pKa = 11.84PNFDD146 pKa = 3.08VSLKK150 pKa = 10.69DD151 pKa = 3.56SQLSDD156 pKa = 3.51SLSIMLNFEE165 pKa = 4.27NLDD168 pKa = 3.67MFAGSVPINISIGLIARR185 pKa = 11.84FYY187 pKa = 9.58NTIDD191 pKa = 3.29PGVRR195 pKa = 11.84IISDD199 pKa = 3.45DD200 pKa = 4.1LNFQGLIGAEE210 pKa = 4.46GISNQDD216 pKa = 3.21MLSNLGDD223 pKa = 3.7MKK225 pKa = 11.29DD226 pKa = 3.58LFSDD230 pKa = 3.68SVIFKK235 pKa = 8.41PTIVSNSKK243 pKa = 10.0FDD245 pKa = 3.51RR246 pKa = 11.84GMFKK250 pKa = 10.68SKK252 pKa = 10.72GLIKK256 pKa = 10.1QIRR259 pKa = 11.84GARR262 pKa = 11.84SGKK265 pKa = 10.03EE266 pKa = 3.57IISTGEE272 pKa = 3.69VKK274 pKa = 10.43RR275 pKa = 11.84PKK277 pKa = 10.22SSSFRR282 pKa = 11.84FDD284 pKa = 3.19QLKK287 pKa = 10.2VDD289 pKa = 3.54PVLYY293 pKa = 10.62DD294 pKa = 3.19KK295 pKa = 11.35LKK297 pKa = 10.43RR298 pKa = 11.84DD299 pKa = 4.69QVNLWLNKK307 pKa = 10.13NNEE310 pKa = 4.06LPIKK314 pKa = 10.27NVEE317 pKa = 4.18DD318 pKa = 3.84EE319 pKa = 5.06EE320 pKa = 4.67IVHH323 pKa = 7.36DD324 pKa = 4.77DD325 pKa = 3.62NSSGRR330 pKa = 11.84VDD332 pKa = 3.17IFEE335 pKa = 4.94HH336 pKa = 6.61GNN338 pKa = 3.16
MM1 pKa = 7.52SLVDD5 pKa = 3.78VNKK8 pKa = 9.99FCSLVSGGKK17 pKa = 9.85SSTEE21 pKa = 4.32AISSSNIYY29 pKa = 10.58GDD31 pKa = 4.31SSWLKK36 pKa = 10.29LKK38 pKa = 10.87NITAIKK44 pKa = 10.2KK45 pKa = 9.69FEE47 pKa = 4.36TNIEE51 pKa = 4.12LEE53 pKa = 4.43LEE55 pKa = 4.77DD56 pKa = 5.11GDD58 pKa = 4.4SKK60 pKa = 11.46LVIPNLPLISDD71 pKa = 3.81SEE73 pKa = 3.9IRR75 pKa = 11.84AVKK78 pKa = 10.61KK79 pKa = 10.63NMPSVNFVHH88 pKa = 6.96LGGIVVSIQALFAANKK104 pKa = 8.49GVKK107 pKa = 8.37GTAVLVDD114 pKa = 3.68RR115 pKa = 11.84RR116 pKa = 11.84WRR118 pKa = 11.84NLDD121 pKa = 3.09QAIIGSFAFNLDD133 pKa = 2.89KK134 pKa = 11.02RR135 pKa = 11.84RR136 pKa = 11.84ADD138 pKa = 3.54FMMRR142 pKa = 11.84PNFDD146 pKa = 3.08VSLKK150 pKa = 10.69DD151 pKa = 3.56SQLSDD156 pKa = 3.51SLSIMLNFEE165 pKa = 4.27NLDD168 pKa = 3.67MFAGSVPINISIGLIARR185 pKa = 11.84FYY187 pKa = 9.58NTIDD191 pKa = 3.29PGVRR195 pKa = 11.84IISDD199 pKa = 3.45DD200 pKa = 4.1LNFQGLIGAEE210 pKa = 4.46GISNQDD216 pKa = 3.21MLSNLGDD223 pKa = 3.7MKK225 pKa = 11.29DD226 pKa = 3.58LFSDD230 pKa = 3.68SVIFKK235 pKa = 8.41PTIVSNSKK243 pKa = 10.0FDD245 pKa = 3.51RR246 pKa = 11.84GMFKK250 pKa = 10.68SKK252 pKa = 10.72GLIKK256 pKa = 10.1QIRR259 pKa = 11.84GARR262 pKa = 11.84SGKK265 pKa = 10.03EE266 pKa = 3.57IISTGEE272 pKa = 3.69VKK274 pKa = 10.43RR275 pKa = 11.84PKK277 pKa = 10.22SSSFRR282 pKa = 11.84FDD284 pKa = 3.19QLKK287 pKa = 10.2VDD289 pKa = 3.54PVLYY293 pKa = 10.62DD294 pKa = 3.19KK295 pKa = 11.35LKK297 pKa = 10.43RR298 pKa = 11.84DD299 pKa = 4.69QVNLWLNKK307 pKa = 10.13NNEE310 pKa = 4.06LPIKK314 pKa = 10.27NVEE317 pKa = 4.18DD318 pKa = 3.84EE319 pKa = 5.06EE320 pKa = 4.67IVHH323 pKa = 7.36DD324 pKa = 4.77DD325 pKa = 3.62NSSGRR330 pKa = 11.84VDD332 pKa = 3.17IFEE335 pKa = 4.94HH336 pKa = 6.61GNN338 pKa = 3.16
Molecular weight: 37.59 kDa
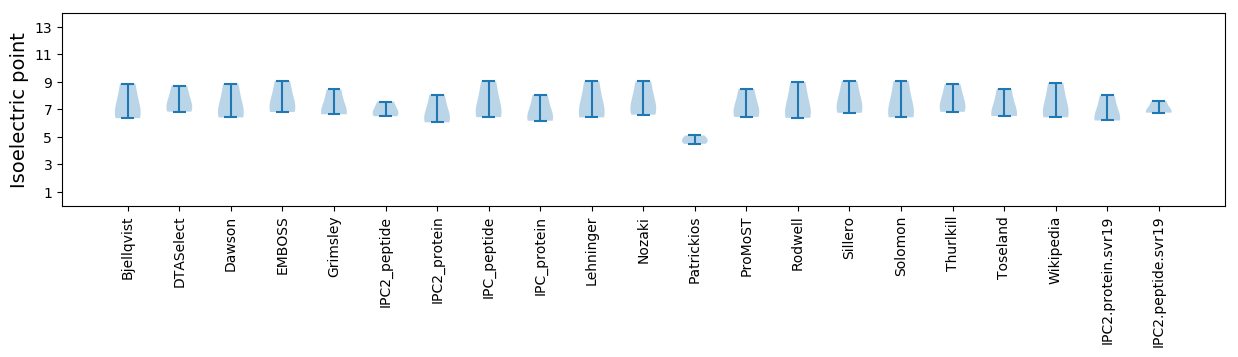
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0P9W1|A0A0A0P9W1_9VIRU Coat protein OS=Carrot Ch virus 1 OX=1425363 PE=4 SV=1
MM1 pKa = 7.28SCRR4 pKa = 11.84SRR6 pKa = 11.84MLKK9 pKa = 8.28TRR11 pKa = 11.84RR12 pKa = 11.84SFMMTILLEE21 pKa = 4.07EE22 pKa = 5.22LISLNTGTEE31 pKa = 3.56IGKK34 pKa = 10.53SIDD37 pKa = 3.56SLDD40 pKa = 4.52LIGNSLLNNYY50 pKa = 8.96RR51 pKa = 11.84DD52 pKa = 3.36HH53 pKa = 7.13FLINMAEE60 pKa = 4.46LGTSPDD66 pKa = 3.18TLYY69 pKa = 11.22EE70 pKa = 4.02MEE72 pKa = 4.78PFEE75 pKa = 4.62LPKK78 pKa = 10.08VTLVSNGEE86 pKa = 4.01KK87 pKa = 9.78KK88 pKa = 10.37DD89 pKa = 3.8YY90 pKa = 9.28KK91 pKa = 10.81LKK93 pKa = 10.36MSIGSAVNSIMNQIEE108 pKa = 4.24GDD110 pKa = 3.82HH111 pKa = 7.02RR112 pKa = 11.84LLTCNTTLRR121 pKa = 11.84NTLGAFGTEE130 pKa = 4.35TKK132 pKa = 10.44RR133 pKa = 11.84QLKK136 pKa = 8.8NRR138 pKa = 11.84KK139 pKa = 8.63IDD141 pKa = 4.18GILARR146 pKa = 11.84KK147 pKa = 8.67YY148 pKa = 10.53PKK150 pKa = 10.0LCKK153 pKa = 9.55QAMHH157 pKa = 6.58MGFDD161 pKa = 4.42FNQDD165 pKa = 2.54IPTNEE170 pKa = 3.79LSKK173 pKa = 11.28SEE175 pKa = 4.18MKK177 pKa = 10.47VKK179 pKa = 10.39QYY181 pKa = 11.14LYY183 pKa = 9.95QQLKK187 pKa = 9.49RR188 pKa = 11.84KK189 pKa = 6.93EE190 pKa = 4.08ARR192 pKa = 11.84RR193 pKa = 11.84GQYY196 pKa = 10.1EE197 pKa = 3.91EE198 pKa = 5.53DD199 pKa = 3.14ISVVDD204 pKa = 5.41FEE206 pKa = 4.7
MM1 pKa = 7.28SCRR4 pKa = 11.84SRR6 pKa = 11.84MLKK9 pKa = 8.28TRR11 pKa = 11.84RR12 pKa = 11.84SFMMTILLEE21 pKa = 4.07EE22 pKa = 5.22LISLNTGTEE31 pKa = 3.56IGKK34 pKa = 10.53SIDD37 pKa = 3.56SLDD40 pKa = 4.52LIGNSLLNNYY50 pKa = 8.96RR51 pKa = 11.84DD52 pKa = 3.36HH53 pKa = 7.13FLINMAEE60 pKa = 4.46LGTSPDD66 pKa = 3.18TLYY69 pKa = 11.22EE70 pKa = 4.02MEE72 pKa = 4.78PFEE75 pKa = 4.62LPKK78 pKa = 10.08VTLVSNGEE86 pKa = 4.01KK87 pKa = 9.78KK88 pKa = 10.37DD89 pKa = 3.8YY90 pKa = 9.28KK91 pKa = 10.81LKK93 pKa = 10.36MSIGSAVNSIMNQIEE108 pKa = 4.24GDD110 pKa = 3.82HH111 pKa = 7.02RR112 pKa = 11.84LLTCNTTLRR121 pKa = 11.84NTLGAFGTEE130 pKa = 4.35TKK132 pKa = 10.44RR133 pKa = 11.84QLKK136 pKa = 8.8NRR138 pKa = 11.84KK139 pKa = 8.63IDD141 pKa = 4.18GILARR146 pKa = 11.84KK147 pKa = 8.67YY148 pKa = 10.53PKK150 pKa = 10.0LCKK153 pKa = 9.55QAMHH157 pKa = 6.58MGFDD161 pKa = 4.42FNQDD165 pKa = 2.54IPTNEE170 pKa = 3.79LSKK173 pKa = 11.28SEE175 pKa = 4.18MKK177 pKa = 10.47VKK179 pKa = 10.39QYY181 pKa = 11.14LYY183 pKa = 9.95QQLKK187 pKa = 9.49RR188 pKa = 11.84KK189 pKa = 6.93EE190 pKa = 4.08ARR192 pKa = 11.84RR193 pKa = 11.84GQYY196 pKa = 10.1EE197 pKa = 3.91EE198 pKa = 5.53DD199 pKa = 3.14ISVVDD204 pKa = 5.41FEE206 pKa = 4.7
Molecular weight: 23.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2401 |
206 |
1857 |
800.3 |
91.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.373 ± 0.453 | 1.208 ± 0.274 |
7.08 ± 0.587 | 6.497 ± 0.647 |
6.039 ± 0.82 | 5.623 ± 0.391 |
1.749 ± 0.306 | 7.455 ± 0.547 |
8.705 ± 0.222 | 10.162 ± 0.557 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.207 ± 0.599 | 5.748 ± 0.675 |
2.874 ± 0.126 | 2.457 ± 0.41 |
4.831 ± 0.425 | 9.08 ± 0.731 |
3.915 ± 0.877 | 5.248 ± 0.761 |
0.916 ± 0.271 | 2.832 ± 0.585 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
