
Mesonia sp. K7
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mesonia; unclassified Mesonia
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
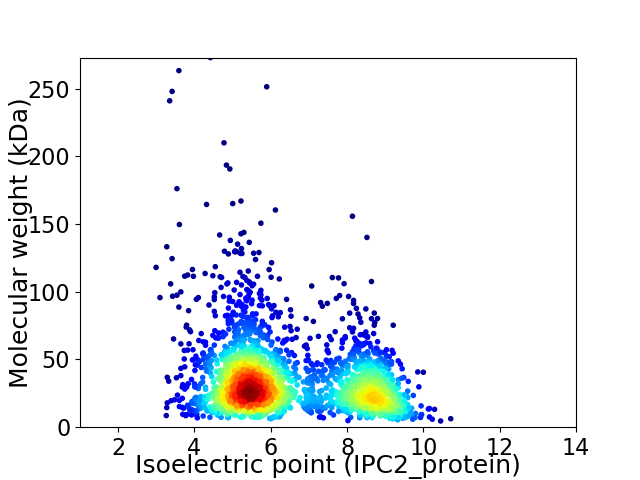
Virtual 2D-PAGE plot for 2375 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W1K6Y6|A0A2W1K6Y6_9FLAO DUF192 domain-containing protein OS=Mesonia sp. K7 OX=2218606 GN=DNG35_06485 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.33KK3 pKa = 10.44LLFFYY8 pKa = 10.56FLLIGFYY15 pKa = 10.11TYY17 pKa = 11.0GQVGIGTAMPDD28 pKa = 3.27PSAQLDD34 pKa = 3.82VTVAAGDD41 pKa = 3.65KK42 pKa = 10.74GVLLPRR48 pKa = 11.84VALINTTDD56 pKa = 3.02QTTITNGNVEE66 pKa = 4.3SLLVYY71 pKa = 9.2NTATQNNVTPGYY83 pKa = 8.59YY84 pKa = 9.55YY85 pKa = 9.88WYY87 pKa = 8.5NGSWRR92 pKa = 11.84RR93 pKa = 11.84LTALGDD99 pKa = 3.69HH100 pKa = 7.42PEE102 pKa = 4.51TITTLVNNTDD112 pKa = 2.62GSYY115 pKa = 10.05TYY117 pKa = 10.1TSEE120 pKa = 4.68DD121 pKa = 2.97GTMTTFTVGGMMVDD135 pKa = 3.82NGDD138 pKa = 3.26GTYY141 pKa = 10.64TFTNPDD147 pKa = 3.3GTNVTIQGVSQALTILSYY165 pKa = 11.63DD166 pKa = 3.71NNTGMISYY174 pKa = 9.76IDD176 pKa = 3.87EE177 pKa = 4.55NSTLTQLDD185 pKa = 3.49MSLAIDD191 pKa = 3.27SWEE194 pKa = 4.0TLTDD198 pKa = 4.08LKK200 pKa = 11.2LNPDD204 pKa = 4.81NIHH207 pKa = 7.18LDD209 pKa = 3.6YY210 pKa = 10.88TDD212 pKa = 3.77EE213 pKa = 5.83DD214 pKa = 4.67GVTNQIDD221 pKa = 4.3LTTLVQNLEE230 pKa = 4.11TLTTIVANADD240 pKa = 3.3GTFTYY245 pKa = 10.53TDD247 pKa = 3.44EE248 pKa = 4.83NGNATQIDD256 pKa = 3.9ISNLEE261 pKa = 4.23TLTSLALNADD271 pKa = 4.08GKK273 pKa = 8.0TLEE276 pKa = 4.29YY277 pKa = 9.66TDD279 pKa = 4.0EE280 pKa = 4.75SGTVTTVDD288 pKa = 3.67LSSVITNFEE297 pKa = 4.54TITTLVDD304 pKa = 3.59NGDD307 pKa = 3.23GTYY310 pKa = 10.07TYY312 pKa = 10.47TNEE315 pKa = 4.26SGNTTVIDD323 pKa = 3.88VNNTEE328 pKa = 4.29TLTEE332 pKa = 4.26LVLSTDD338 pKa = 3.41NSTLTYY344 pKa = 10.04TDD346 pKa = 3.61EE347 pKa = 4.64NGNDD351 pKa = 3.63TIINLKK357 pKa = 9.07TVVQANQKK365 pKa = 6.03TTTLADD371 pKa = 3.76GVNTTVSSTVSGDD384 pKa = 3.1NTAWQVNVATAKK396 pKa = 10.34GANNTQNSTLGVVKK410 pKa = 10.15EE411 pKa = 4.14ADD413 pKa = 3.58SNPTVNIANDD423 pKa = 3.66GSLSVNLEE431 pKa = 3.62NTNDD435 pKa = 3.3IKK437 pKa = 10.96EE438 pKa = 4.04VSGNYY443 pKa = 8.2TVLPNDD449 pKa = 4.2TILLGNASTADD460 pKa = 3.53VSIVMPNPVGKK471 pKa = 9.99KK472 pKa = 10.26GKK474 pKa = 8.71TITIKK479 pKa = 10.81KK480 pKa = 9.56QDD482 pKa = 3.8TNEE485 pKa = 3.75DD486 pKa = 4.14YY487 pKa = 11.3YY488 pKa = 10.83INVYY492 pKa = 10.98GNINGLSQLYY502 pKa = 7.91TALPYY507 pKa = 10.78SGWQLTSDD515 pKa = 3.84GTQWKK520 pKa = 7.93ITNKK524 pKa = 9.86FF525 pKa = 3.15
MM1 pKa = 7.5KK2 pKa = 10.33KK3 pKa = 10.44LLFFYY8 pKa = 10.56FLLIGFYY15 pKa = 10.11TYY17 pKa = 11.0GQVGIGTAMPDD28 pKa = 3.27PSAQLDD34 pKa = 3.82VTVAAGDD41 pKa = 3.65KK42 pKa = 10.74GVLLPRR48 pKa = 11.84VALINTTDD56 pKa = 3.02QTTITNGNVEE66 pKa = 4.3SLLVYY71 pKa = 9.2NTATQNNVTPGYY83 pKa = 8.59YY84 pKa = 9.55YY85 pKa = 9.88WYY87 pKa = 8.5NGSWRR92 pKa = 11.84RR93 pKa = 11.84LTALGDD99 pKa = 3.69HH100 pKa = 7.42PEE102 pKa = 4.51TITTLVNNTDD112 pKa = 2.62GSYY115 pKa = 10.05TYY117 pKa = 10.1TSEE120 pKa = 4.68DD121 pKa = 2.97GTMTTFTVGGMMVDD135 pKa = 3.82NGDD138 pKa = 3.26GTYY141 pKa = 10.64TFTNPDD147 pKa = 3.3GTNVTIQGVSQALTILSYY165 pKa = 11.63DD166 pKa = 3.71NNTGMISYY174 pKa = 9.76IDD176 pKa = 3.87EE177 pKa = 4.55NSTLTQLDD185 pKa = 3.49MSLAIDD191 pKa = 3.27SWEE194 pKa = 4.0TLTDD198 pKa = 4.08LKK200 pKa = 11.2LNPDD204 pKa = 4.81NIHH207 pKa = 7.18LDD209 pKa = 3.6YY210 pKa = 10.88TDD212 pKa = 3.77EE213 pKa = 5.83DD214 pKa = 4.67GVTNQIDD221 pKa = 4.3LTTLVQNLEE230 pKa = 4.11TLTTIVANADD240 pKa = 3.3GTFTYY245 pKa = 10.53TDD247 pKa = 3.44EE248 pKa = 4.83NGNATQIDD256 pKa = 3.9ISNLEE261 pKa = 4.23TLTSLALNADD271 pKa = 4.08GKK273 pKa = 8.0TLEE276 pKa = 4.29YY277 pKa = 9.66TDD279 pKa = 4.0EE280 pKa = 4.75SGTVTTVDD288 pKa = 3.67LSSVITNFEE297 pKa = 4.54TITTLVDD304 pKa = 3.59NGDD307 pKa = 3.23GTYY310 pKa = 10.07TYY312 pKa = 10.47TNEE315 pKa = 4.26SGNTTVIDD323 pKa = 3.88VNNTEE328 pKa = 4.29TLTEE332 pKa = 4.26LVLSTDD338 pKa = 3.41NSTLTYY344 pKa = 10.04TDD346 pKa = 3.61EE347 pKa = 4.64NGNDD351 pKa = 3.63TIINLKK357 pKa = 9.07TVVQANQKK365 pKa = 6.03TTTLADD371 pKa = 3.76GVNTTVSSTVSGDD384 pKa = 3.1NTAWQVNVATAKK396 pKa = 10.34GANNTQNSTLGVVKK410 pKa = 10.15EE411 pKa = 4.14ADD413 pKa = 3.58SNPTVNIANDD423 pKa = 3.66GSLSVNLEE431 pKa = 3.62NTNDD435 pKa = 3.3IKK437 pKa = 10.96EE438 pKa = 4.04VSGNYY443 pKa = 8.2TVLPNDD449 pKa = 4.2TILLGNASTADD460 pKa = 3.53VSIVMPNPVGKK471 pKa = 9.99KK472 pKa = 10.26GKK474 pKa = 8.71TITIKK479 pKa = 10.81KK480 pKa = 9.56QDD482 pKa = 3.8TNEE485 pKa = 3.75DD486 pKa = 4.14YY487 pKa = 11.3YY488 pKa = 10.83INVYY492 pKa = 10.98GNINGLSQLYY502 pKa = 7.91TALPYY507 pKa = 10.78SGWQLTSDD515 pKa = 3.84GTQWKK520 pKa = 7.93ITNKK524 pKa = 9.86FF525 pKa = 3.15
Molecular weight: 56.68 kDa
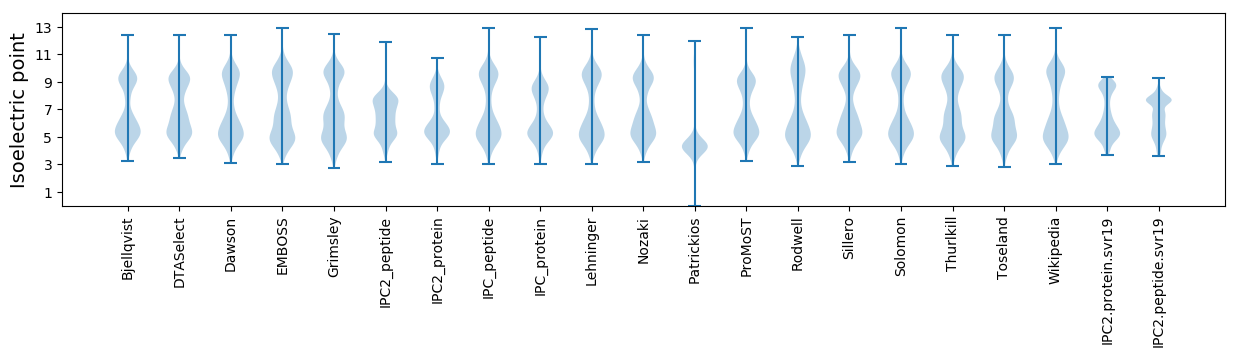
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W1KG00|A0A2W1KG00_9FLAO Uncharacterized protein OS=Mesonia sp. K7 OX=2218606 GN=DNG35_05730 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.01RR21 pKa = 11.84MATVNGRR28 pKa = 11.84KK29 pKa = 9.18VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.01RR21 pKa = 11.84MATVNGRR28 pKa = 11.84KK29 pKa = 9.18VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
780029 |
38 |
2477 |
328.4 |
37.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.245 ± 0.049 | 0.72 ± 0.015 |
5.429 ± 0.04 | 7.275 ± 0.058 |
5.234 ± 0.046 | 6.076 ± 0.055 |
1.819 ± 0.024 | 8.0 ± 0.048 |
8.027 ± 0.09 | 9.16 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.126 ± 0.026 | 6.214 ± 0.056 |
3.333 ± 0.035 | 3.874 ± 0.033 |
3.295 ± 0.032 | 6.104 ± 0.042 |
5.726 ± 0.074 | 6.194 ± 0.036 |
0.961 ± 0.02 | 4.189 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
