
Undibacterium pigrum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Undibacterium
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
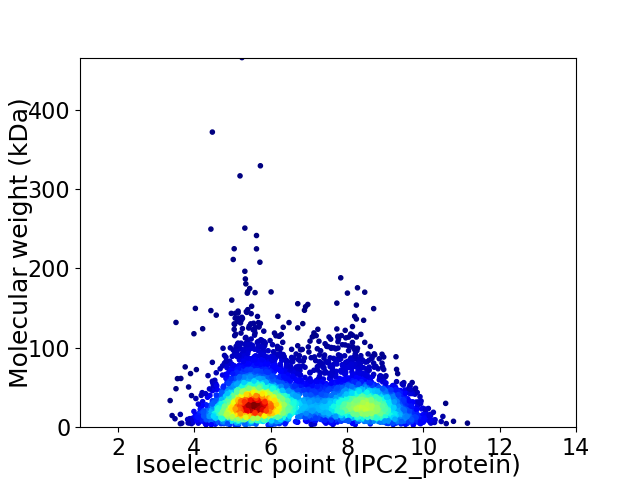
Virtual 2D-PAGE plot for 5704 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A318JBN9|A0A318JBN9_9BURK Amino acid/amide ABC transporter substrate-binding protein (HAAT family) OS=Undibacterium pigrum OX=401470 GN=DFR42_102218 PE=4 SV=1
TT1 pKa = 7.14YY2 pKa = 7.63NTTEE6 pKa = 3.92NGVNLNAQSTANAVARR22 pKa = 11.84VVTDD26 pKa = 4.23TINLGAGNDD35 pKa = 3.74TLVTYY40 pKa = 9.55GAINLAGAQLSNIEE54 pKa = 4.63NITSNSAVVITASQYY69 pKa = 10.69AALIAARR76 pKa = 11.84AALNLSGPVLTFSGVGPHH94 pKa = 6.06QLTIVDD100 pKa = 4.56DD101 pKa = 3.98VAGANNIDD109 pKa = 3.98LSFISITGGTLVYY122 pKa = 10.69DD123 pKa = 3.89VTSSSNATGGGVNNTTTSGAVTVSGSGVAIAGTIGTSPSNGGGQAANGTALTAGGTFNGTAGLNEE188 pKa = 4.38NFTGNAAALVGTTVNGNNGDD208 pKa = 3.7TDD210 pKa = 4.43TITISGGGTVNIGSGNTITNIKK232 pKa = 8.21TLTMDD237 pKa = 3.62SAAANVVFFSVANSGITTVNGGSGNDD263 pKa = 3.66YY264 pKa = 11.26VDD266 pKa = 4.34LANSGNSMLAGNVNLGTGTNTLVMEE291 pKa = 5.0GKK293 pKa = 8.63TYY295 pKa = 10.45TGTFVSGSGTSDD307 pKa = 2.84TLYY310 pKa = 10.92LFNGSNISAANVSGFEE326 pKa = 4.14TVNINNNASVTMTAAQFAGFTTFTAGGTEE355 pKa = 4.37TVNLTTAGTVTANANVEE372 pKa = 4.16NYY374 pKa = 10.01VLANGSNTFTGAAGLISVNGGTGSDD399 pKa = 4.01TINVSSVMISATVAPNGINGGLGADD424 pKa = 3.94TLNVSAVTAALDD436 pKa = 3.48MSAKK440 pKa = 8.0VTGVEE445 pKa = 4.38TVNVTGGTNAAWTVTNEE462 pKa = 3.64NGAGVTLNFTKK473 pKa = 10.67SAANDD478 pKa = 3.42INNITLGSGGQTLNILGTGTGRR500 pKa = 11.84TTITGGSGADD510 pKa = 3.77IINLSATATGADD522 pKa = 4.14AIALGSNINAIDD534 pKa = 3.54TVNNFKK540 pKa = 10.83AAGADD545 pKa = 3.71LFATGVVPTSLNNLSIANADD565 pKa = 3.76SSNLAAAIATAATAAGATLANTGQAYY591 pKa = 8.61TIVVNSGTAAGIYY604 pKa = 10.02AFQNTGGSVGAVDD617 pKa = 3.37TGDD620 pKa = 4.34FIVKK624 pKa = 10.53LGGTTGTIFATDD636 pKa = 3.68FGIANAVAVTPGGTFNGTAGANDD659 pKa = 3.47IFMSTIPGLNGTTIAGNAADD679 pKa = 4.34TDD681 pKa = 4.16VLTLTTAGTVTINNGSTGGTLSNLKK706 pKa = 9.93VLNLANGTNTITYY719 pKa = 7.22NTSAGFTTINGGSGDD734 pKa = 3.91DD735 pKa = 3.47TFIPNTALLPIVVSGGLGTDD755 pKa = 4.11TIVLSAAYY763 pKa = 9.47AAVASGSGTFAGNVTGFEE781 pKa = 4.29KK782 pKa = 10.84LRR784 pKa = 11.84LTSVTNQTIDD794 pKa = 3.29LQTLGNYY801 pKa = 10.24SDD803 pKa = 3.85VTFSGANGLTMSNLPSNGNITLNGTGTAFTISNAAFAGGVNDD845 pKa = 4.4IVNLTLTDD853 pKa = 3.69ASTSAVAFATTGITASGVEE872 pKa = 4.37TFNITTVDD880 pKa = 3.62GQATPTGLFNDD891 pKa = 3.99TLTILGNSVKK901 pKa = 10.56NIYY904 pKa = 10.29VSGNAGLTMPATSTSLINVDD924 pKa = 3.48ASGITLGGFTWTANALTSTATVKK947 pKa = 10.78GSASGTNTVNMNSATAGVDD966 pKa = 3.54YY967 pKa = 10.98TGGTGNDD974 pKa = 3.57NITINATVSSTAALGGGNNSLALNGVTLLGTYY1006 pKa = 7.55TAGNTGTDD1014 pKa = 3.04SLAFFSSTPDD1024 pKa = 2.96ISNATITGFEE1034 pKa = 4.22NLTVVNNANITMTAAQMAQFTGTVNASGTEE1064 pKa = 4.29TINLTTAGTVTAFSAVEE1081 pKa = 4.26KK1082 pKa = 11.03YY1083 pKa = 10.95NLANGTNNFTSANVAVSVIGGTGTDD1108 pKa = 3.41TFNFTANQIANFLTTVDD1125 pKa = 4.43GGNGTDD1131 pKa = 3.59TLNVGATTTQNLDD1144 pKa = 3.24FSTKK1148 pKa = 9.12IASIEE1153 pKa = 4.08IINVAGSTGTASVTNPDD1170 pKa = 3.54GAGVTLNYY1178 pKa = 8.62TKK1180 pKa = 10.01STGDD1184 pKa = 3.23NTITLGTGGQTLNLLGSSASSTTVTGGAAVDD1215 pKa = 4.26TINLQFSGSGSEE1227 pKa = 4.13TLIEE1231 pKa = 4.18TGANMSNRR1239 pKa = 11.84TQVDD1243 pKa = 2.88IVGNFNSTGTDD1254 pKa = 3.49YY1255 pKa = 11.36FKK1257 pKa = 10.72TGVNAVSVGSYY1268 pKa = 10.24IIGNADD1274 pKa = 3.2TGNYY1278 pKa = 7.23LTTIASGLSIVLNNTGQSYY1297 pKa = 10.7LITIQTGTAAGTYY1310 pKa = 9.74LFQNTGSDD1318 pKa = 3.12TSQFDD1323 pKa = 3.3NTDD1326 pKa = 3.63FFVQLTGTIGGIGAGNLIAA1345 pKa = 5.87
TT1 pKa = 7.14YY2 pKa = 7.63NTTEE6 pKa = 3.92NGVNLNAQSTANAVARR22 pKa = 11.84VVTDD26 pKa = 4.23TINLGAGNDD35 pKa = 3.74TLVTYY40 pKa = 9.55GAINLAGAQLSNIEE54 pKa = 4.63NITSNSAVVITASQYY69 pKa = 10.69AALIAARR76 pKa = 11.84AALNLSGPVLTFSGVGPHH94 pKa = 6.06QLTIVDD100 pKa = 4.56DD101 pKa = 3.98VAGANNIDD109 pKa = 3.98LSFISITGGTLVYY122 pKa = 10.69DD123 pKa = 3.89VTSSSNATGGGVNNTTTSGAVTVSGSGVAIAGTIGTSPSNGGGQAANGTALTAGGTFNGTAGLNEE188 pKa = 4.38NFTGNAAALVGTTVNGNNGDD208 pKa = 3.7TDD210 pKa = 4.43TITISGGGTVNIGSGNTITNIKK232 pKa = 8.21TLTMDD237 pKa = 3.62SAAANVVFFSVANSGITTVNGGSGNDD263 pKa = 3.66YY264 pKa = 11.26VDD266 pKa = 4.34LANSGNSMLAGNVNLGTGTNTLVMEE291 pKa = 5.0GKK293 pKa = 8.63TYY295 pKa = 10.45TGTFVSGSGTSDD307 pKa = 2.84TLYY310 pKa = 10.92LFNGSNISAANVSGFEE326 pKa = 4.14TVNINNNASVTMTAAQFAGFTTFTAGGTEE355 pKa = 4.37TVNLTTAGTVTANANVEE372 pKa = 4.16NYY374 pKa = 10.01VLANGSNTFTGAAGLISVNGGTGSDD399 pKa = 4.01TINVSSVMISATVAPNGINGGLGADD424 pKa = 3.94TLNVSAVTAALDD436 pKa = 3.48MSAKK440 pKa = 8.0VTGVEE445 pKa = 4.38TVNVTGGTNAAWTVTNEE462 pKa = 3.64NGAGVTLNFTKK473 pKa = 10.67SAANDD478 pKa = 3.42INNITLGSGGQTLNILGTGTGRR500 pKa = 11.84TTITGGSGADD510 pKa = 3.77IINLSATATGADD522 pKa = 4.14AIALGSNINAIDD534 pKa = 3.54TVNNFKK540 pKa = 10.83AAGADD545 pKa = 3.71LFATGVVPTSLNNLSIANADD565 pKa = 3.76SSNLAAAIATAATAAGATLANTGQAYY591 pKa = 8.61TIVVNSGTAAGIYY604 pKa = 10.02AFQNTGGSVGAVDD617 pKa = 3.37TGDD620 pKa = 4.34FIVKK624 pKa = 10.53LGGTTGTIFATDD636 pKa = 3.68FGIANAVAVTPGGTFNGTAGANDD659 pKa = 3.47IFMSTIPGLNGTTIAGNAADD679 pKa = 4.34TDD681 pKa = 4.16VLTLTTAGTVTINNGSTGGTLSNLKK706 pKa = 9.93VLNLANGTNTITYY719 pKa = 7.22NTSAGFTTINGGSGDD734 pKa = 3.91DD735 pKa = 3.47TFIPNTALLPIVVSGGLGTDD755 pKa = 4.11TIVLSAAYY763 pKa = 9.47AAVASGSGTFAGNVTGFEE781 pKa = 4.29KK782 pKa = 10.84LRR784 pKa = 11.84LTSVTNQTIDD794 pKa = 3.29LQTLGNYY801 pKa = 10.24SDD803 pKa = 3.85VTFSGANGLTMSNLPSNGNITLNGTGTAFTISNAAFAGGVNDD845 pKa = 4.4IVNLTLTDD853 pKa = 3.69ASTSAVAFATTGITASGVEE872 pKa = 4.37TFNITTVDD880 pKa = 3.62GQATPTGLFNDD891 pKa = 3.99TLTILGNSVKK901 pKa = 10.56NIYY904 pKa = 10.29VSGNAGLTMPATSTSLINVDD924 pKa = 3.48ASGITLGGFTWTANALTSTATVKK947 pKa = 10.78GSASGTNTVNMNSATAGVDD966 pKa = 3.54YY967 pKa = 10.98TGGTGNDD974 pKa = 3.57NITINATVSSTAALGGGNNSLALNGVTLLGTYY1006 pKa = 7.55TAGNTGTDD1014 pKa = 3.04SLAFFSSTPDD1024 pKa = 2.96ISNATITGFEE1034 pKa = 4.22NLTVVNNANITMTAAQMAQFTGTVNASGTEE1064 pKa = 4.29TINLTTAGTVTAFSAVEE1081 pKa = 4.26KK1082 pKa = 11.03YY1083 pKa = 10.95NLANGTNNFTSANVAVSVIGGTGTDD1108 pKa = 3.41TFNFTANQIANFLTTVDD1125 pKa = 4.43GGNGTDD1131 pKa = 3.59TLNVGATTTQNLDD1144 pKa = 3.24FSTKK1148 pKa = 9.12IASIEE1153 pKa = 4.08IINVAGSTGTASVTNPDD1170 pKa = 3.54GAGVTLNYY1178 pKa = 8.62TKK1180 pKa = 10.01STGDD1184 pKa = 3.23NTITLGTGGQTLNLLGSSASSTTVTGGAAVDD1215 pKa = 4.26TINLQFSGSGSEE1227 pKa = 4.13TLIEE1231 pKa = 4.18TGANMSNRR1239 pKa = 11.84TQVDD1243 pKa = 2.88IVGNFNSTGTDD1254 pKa = 3.49YY1255 pKa = 11.36FKK1257 pKa = 10.72TGVNAVSVGSYY1268 pKa = 10.24IIGNADD1274 pKa = 3.2TGNYY1278 pKa = 7.23LTTIASGLSIVLNNTGQSYY1297 pKa = 10.7LITIQTGTAAGTYY1310 pKa = 9.74LFQNTGSDD1318 pKa = 3.12TSQFDD1323 pKa = 3.3NTDD1326 pKa = 3.63FFVQLTGTIGGIGAGNLIAA1345 pKa = 5.87
Molecular weight: 131.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A318JMY3|A0A318JMY3_9BURK Uncharacterized protein OS=Undibacterium pigrum OX=401470 GN=DFR42_102579 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84AVLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84AVLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1883445 |
27 |
4271 |
330.2 |
36.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.924 ± 0.044 | 0.938 ± 0.011 |
5.266 ± 0.026 | 5.41 ± 0.034 |
3.883 ± 0.023 | 7.163 ± 0.035 |
2.23 ± 0.018 | 5.638 ± 0.024 |
4.798 ± 0.031 | 10.609 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.701 ± 0.015 | 3.748 ± 0.024 |
4.37 ± 0.023 | 4.534 ± 0.03 |
5.253 ± 0.024 | 6.339 ± 0.026 |
5.343 ± 0.031 | 6.735 ± 0.031 |
1.343 ± 0.014 | 2.773 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
