
Senegalimassilia anaerobia
Taxonomy: cellular organisms; Bacteria;
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
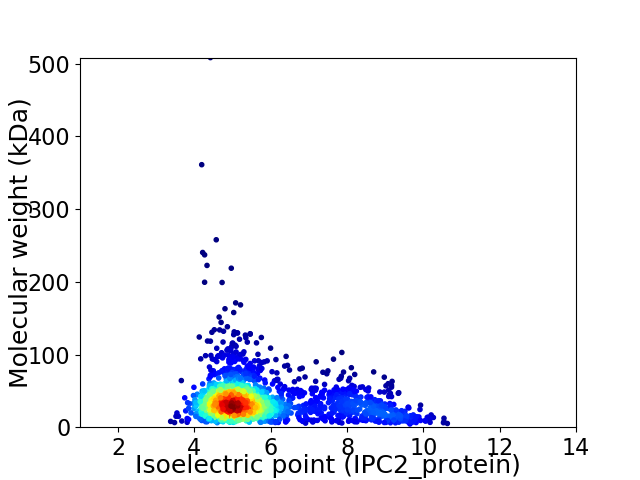
Virtual 2D-PAGE plot for 1843 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369LCJ5|A0A369LCJ5_9ACTN Undecaprenyl-diphosphatase OS=Senegalimassilia anaerobia OX=1473216 GN=uppP PE=3 SV=1
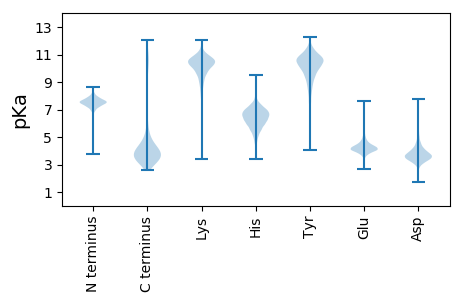
MM1 pKa = 7.51KK2 pKa = 10.37SYY4 pKa = 10.8IEE6 pKa = 4.32PLNAADD12 pKa = 5.21ALDD15 pKa = 3.7VLDD18 pKa = 5.74LYY20 pKa = 11.56GIDD23 pKa = 3.43TPGAFFSSEE32 pKa = 3.96HH33 pKa = 5.9EE34 pKa = 4.33GCVEE38 pKa = 4.03VEE40 pKa = 4.27LSGHH44 pKa = 5.04TALLEE49 pKa = 4.36TPEE52 pKa = 4.17SVAMLFGHH60 pKa = 7.42DD61 pKa = 4.21AATALCQHH69 pKa = 6.27SNFRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.01AAEE78 pKa = 3.8IMSRR82 pKa = 11.84ALDD85 pKa = 4.23LDD87 pKa = 3.96DD88 pKa = 5.35FLYY91 pKa = 10.93NVYY94 pKa = 10.48SGTAFIPDD102 pKa = 3.83KK103 pKa = 11.31APDD106 pKa = 3.65GEE108 pKa = 4.42FFGFYY113 pKa = 9.34VAHH116 pKa = 6.91IPLSAILEE124 pKa = 4.3DD125 pKa = 6.15DD126 pKa = 4.37IDD128 pKa = 4.31PLSPDD133 pKa = 3.06ACDD136 pKa = 3.65YY137 pKa = 11.37GLDD140 pKa = 4.19SISEE144 pKa = 4.34TTEE147 pKa = 3.86DD148 pKa = 3.39AAVGYY153 pKa = 8.96FDD155 pKa = 4.84PAGAVDD161 pKa = 4.72DD162 pKa = 4.89EE163 pKa = 4.85KK164 pKa = 11.33FLTSADD170 pKa = 3.22WALCDD175 pKa = 3.99IEE177 pKa = 5.94SSVLTAFGAGCSPEE191 pKa = 4.0LAGWTLGNAEE201 pKa = 4.75SVAEE205 pKa = 4.37TCASLGVDD213 pKa = 3.37IATAPAPDD221 pKa = 3.5NPARR225 pKa = 11.84VAAAAKK231 pKa = 9.87AATKK235 pKa = 10.37AATHH239 pKa = 6.17GVAAAAGNRR248 pKa = 11.84LL249 pKa = 3.39
MM1 pKa = 7.51KK2 pKa = 10.37SYY4 pKa = 10.8IEE6 pKa = 4.32PLNAADD12 pKa = 5.21ALDD15 pKa = 3.7VLDD18 pKa = 5.74LYY20 pKa = 11.56GIDD23 pKa = 3.43TPGAFFSSEE32 pKa = 3.96HH33 pKa = 5.9EE34 pKa = 4.33GCVEE38 pKa = 4.03VEE40 pKa = 4.27LSGHH44 pKa = 5.04TALLEE49 pKa = 4.36TPEE52 pKa = 4.17SVAMLFGHH60 pKa = 7.42DD61 pKa = 4.21AATALCQHH69 pKa = 6.27SNFRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.01AAEE78 pKa = 3.8IMSRR82 pKa = 11.84ALDD85 pKa = 4.23LDD87 pKa = 3.96DD88 pKa = 5.35FLYY91 pKa = 10.93NVYY94 pKa = 10.48SGTAFIPDD102 pKa = 3.83KK103 pKa = 11.31APDD106 pKa = 3.65GEE108 pKa = 4.42FFGFYY113 pKa = 9.34VAHH116 pKa = 6.91IPLSAILEE124 pKa = 4.3DD125 pKa = 6.15DD126 pKa = 4.37IDD128 pKa = 4.31PLSPDD133 pKa = 3.06ACDD136 pKa = 3.65YY137 pKa = 11.37GLDD140 pKa = 4.19SISEE144 pKa = 4.34TTEE147 pKa = 3.86DD148 pKa = 3.39AAVGYY153 pKa = 8.96FDD155 pKa = 4.84PAGAVDD161 pKa = 4.72DD162 pKa = 4.89EE163 pKa = 4.85KK164 pKa = 11.33FLTSADD170 pKa = 3.22WALCDD175 pKa = 3.99IEE177 pKa = 5.94SSVLTAFGAGCSPEE191 pKa = 4.0LAGWTLGNAEE201 pKa = 4.75SVAEE205 pKa = 4.37TCASLGVDD213 pKa = 3.37IATAPAPDD221 pKa = 3.5NPARR225 pKa = 11.84VAAAAKK231 pKa = 9.87AATKK235 pKa = 10.37AATHH239 pKa = 6.17GVAAAAGNRR248 pKa = 11.84LL249 pKa = 3.39
Molecular weight: 25.93 kDa
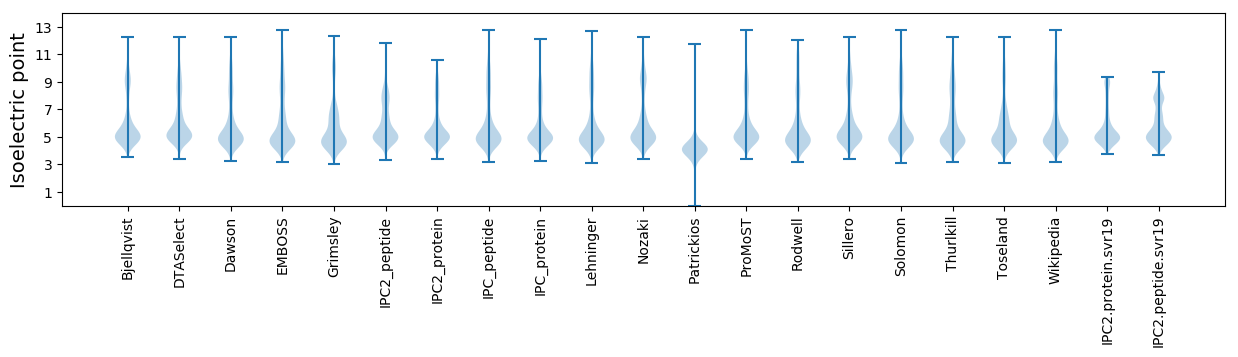
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369L275|A0A369L275_9ACTN Energy-coupling factor transporter transmembrane protein EcfT OS=Senegalimassilia anaerobia OX=1473216 GN=C1880_09410 PE=4 SV=1
MM1 pKa = 7.36SVNGDD6 pKa = 3.37AARR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.33MWAGRR16 pKa = 11.84IEE18 pKa = 4.21RR19 pKa = 11.84CLAADD24 pKa = 3.44MTVKK28 pKa = 10.17EE29 pKa = 4.26WCALNKK35 pKa = 9.9VAEE38 pKa = 4.18SSLYY42 pKa = 10.22KK43 pKa = 9.62WMARR47 pKa = 11.84FRR49 pKa = 11.84EE50 pKa = 4.32EE51 pKa = 3.75EE52 pKa = 4.13PRR54 pKa = 11.84PFPCRR59 pKa = 11.84SSEE62 pKa = 3.77ASSWMRR68 pKa = 11.84VARR71 pKa = 11.84RR72 pKa = 11.84GPGRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84CKK80 pKa = 9.11PVLRR84 pKa = 11.84SRR86 pKa = 11.84YY87 pKa = 8.79GG88 pKa = 3.06
MM1 pKa = 7.36SVNGDD6 pKa = 3.37AARR9 pKa = 11.84RR10 pKa = 11.84DD11 pKa = 3.33MWAGRR16 pKa = 11.84IEE18 pKa = 4.21RR19 pKa = 11.84CLAADD24 pKa = 3.44MTVKK28 pKa = 10.17EE29 pKa = 4.26WCALNKK35 pKa = 9.9VAEE38 pKa = 4.18SSLYY42 pKa = 10.22KK43 pKa = 9.62WMARR47 pKa = 11.84FRR49 pKa = 11.84EE50 pKa = 4.32EE51 pKa = 3.75EE52 pKa = 4.13PRR54 pKa = 11.84PFPCRR59 pKa = 11.84SSEE62 pKa = 3.77ASSWMRR68 pKa = 11.84VARR71 pKa = 11.84RR72 pKa = 11.84GPGRR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84CKK80 pKa = 9.11PVLRR84 pKa = 11.84SRR86 pKa = 11.84YY87 pKa = 8.79GG88 pKa = 3.06
Molecular weight: 10.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
646842 |
37 |
4846 |
351.0 |
38.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.086 ± 0.085 | 1.692 ± 0.032 |
6.18 ± 0.057 | 6.311 ± 0.062 |
3.723 ± 0.04 | 8.356 ± 0.051 |
1.853 ± 0.028 | 4.872 ± 0.051 |
4.004 ± 0.052 | 8.803 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.719 ± 0.036 | 3.016 ± 0.038 |
4.35 ± 0.039 | 3.378 ± 0.036 |
5.76 ± 0.065 | 5.833 ± 0.058 |
5.23 ± 0.058 | 8.023 ± 0.053 |
1.059 ± 0.023 | 2.751 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
