
Reinekea marinisedimentorum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Saccharospirillaceae; Reinekea
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
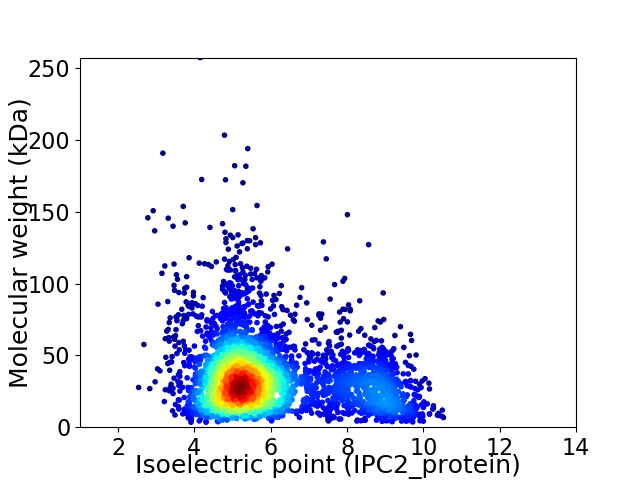
Virtual 2D-PAGE plot for 3800 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R3HU06|A0A4R3HU06_9GAMM Peptide chain release factor 3 OS=Reinekea marinisedimentorum OX=230495 GN=prfC PE=3 SV=1
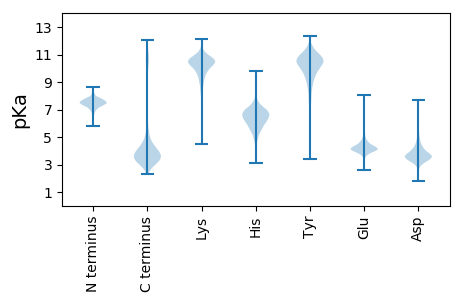
MM1 pKa = 8.05PLTQFKK7 pKa = 10.49RR8 pKa = 11.84KK9 pKa = 9.24ALVVALVSQAGLTATNVAVANDD31 pKa = 3.58STEE34 pKa = 4.02IEE36 pKa = 4.11TDD38 pKa = 3.52TIVISGEE45 pKa = 3.86KK46 pKa = 9.38IEE48 pKa = 5.03RR49 pKa = 11.84SLKK52 pKa = 9.75EE53 pKa = 3.42ATTAVTVITEE63 pKa = 4.36DD64 pKa = 3.31QLQTTEE70 pKa = 4.01TQTINEE76 pKa = 4.38IAASTPNVINGGFGAVSIRR95 pKa = 11.84GVDD98 pKa = 3.48GTGAATGGYY107 pKa = 9.96AFYY110 pKa = 10.53SGARR114 pKa = 11.84ARR116 pKa = 11.84VSTIVDD122 pKa = 3.64GVSQSWSGYY131 pKa = 9.2NFTPSKK137 pKa = 10.44SWDD140 pKa = 3.41VKK142 pKa = 9.5QVEE145 pKa = 5.28VYY147 pKa = 10.44RR148 pKa = 11.84GPQSTTQGTNSIGGAMVVEE167 pKa = 4.96SNDD170 pKa = 3.03PTYY173 pKa = 10.57YY174 pKa = 10.2QEE176 pKa = 3.76AAIRR180 pKa = 11.84LGTEE184 pKa = 3.92VYY186 pKa = 10.79EE187 pKa = 4.69NGNTLSNLAVMASGPLIEE205 pKa = 5.86DD206 pKa = 3.21EE207 pKa = 4.39LAFRR211 pKa = 11.84LAVDD215 pKa = 4.33GSTGEE220 pKa = 4.12SYY222 pKa = 8.67ITYY225 pKa = 9.0EE226 pKa = 4.01QAATEE231 pKa = 4.59LDD233 pKa = 4.01DD234 pKa = 6.71SPDD237 pKa = 3.62LDD239 pKa = 3.46NTTNLNLRR247 pKa = 11.84SKK249 pKa = 10.49LLWEE253 pKa = 4.77PGFLPEE259 pKa = 5.01LSAKK263 pKa = 9.37ATVNYY268 pKa = 10.59SSFDD272 pKa = 3.29GSYY275 pKa = 11.06LNWANDD281 pKa = 3.19TDD283 pKa = 4.24EE284 pKa = 5.95DD285 pKa = 5.17YY286 pKa = 10.78STQTFTVSSADD297 pKa = 3.45RR298 pKa = 11.84DD299 pKa = 3.67YY300 pKa = 11.81TRR302 pKa = 11.84IQDD305 pKa = 3.72STVKK309 pKa = 10.51SVAGDD314 pKa = 3.46ADD316 pKa = 3.91YY317 pKa = 11.52QLTDD321 pKa = 3.38QISNSFQIVYY331 pKa = 10.81AEE333 pKa = 3.87TDD335 pKa = 3.56VEE337 pKa = 4.45FNEE340 pKa = 4.54YY341 pKa = 9.42TSSRR345 pKa = 11.84TVISDD350 pKa = 3.15VDD352 pKa = 3.58NVTVEE357 pKa = 3.51NRR359 pKa = 11.84LNFRR363 pKa = 11.84ASDD366 pKa = 3.7DD367 pKa = 3.65SATAFVGVYY376 pKa = 10.33LSNTEE381 pKa = 3.87TTQDD385 pKa = 3.12IFSVIDD391 pKa = 3.44VEE393 pKa = 5.21SDD395 pKa = 3.28VTTAAVFGEE404 pKa = 4.34ASYY407 pKa = 11.37QLLDD411 pKa = 4.07EE412 pKa = 5.12LSLIAGLRR420 pKa = 11.84YY421 pKa = 10.0EE422 pKa = 4.5NEE424 pKa = 4.01STDD427 pKa = 3.67RR428 pKa = 11.84MLDD431 pKa = 3.2HH432 pKa = 7.6ASNTNYY438 pKa = 10.33NFDD441 pKa = 3.62EE442 pKa = 4.52SVSEE446 pKa = 4.95DD447 pKa = 3.38VLLPKK452 pKa = 10.16IAAIYY457 pKa = 9.65EE458 pKa = 4.28LTDD461 pKa = 3.32NTVVSASVRR470 pKa = 11.84KK471 pKa = 9.91GYY473 pKa = 9.88NAGGGAVNWASDD485 pKa = 3.56TDD487 pKa = 3.93NIGYY491 pKa = 7.58YY492 pKa = 9.68TYY494 pKa = 11.1DD495 pKa = 3.53SEE497 pKa = 5.01TVWAYY502 pKa = 9.62EE503 pKa = 3.95AALKK507 pKa = 10.1TSIAGANVAVSAFINDD523 pKa = 3.51YY524 pKa = 11.33ADD526 pKa = 3.31YY527 pKa = 10.49QAFVSEE533 pKa = 4.57YY534 pKa = 10.77DD535 pKa = 3.35SSLDD539 pKa = 3.42DD540 pKa = 3.32TLQYY544 pKa = 10.44IEE546 pKa = 4.59NVEE549 pKa = 4.1DD550 pKa = 3.63ALTYY554 pKa = 10.36GVEE557 pKa = 4.1LEE559 pKa = 4.02GDD561 pKa = 3.09KK562 pKa = 10.67WLTDD566 pKa = 3.63STSLRR571 pKa = 11.84ASVGYY576 pKa = 7.78TQTEE580 pKa = 4.65VISDD584 pKa = 4.14DD585 pKa = 3.6DD586 pKa = 4.48TIDD589 pKa = 4.25GNEE592 pKa = 4.61LPNAPAFNTSVGVTQYY608 pKa = 10.78FSDD611 pKa = 4.01RR612 pKa = 11.84FSVGADD618 pKa = 2.97VVYY621 pKa = 10.84VGEE624 pKa = 4.51YY625 pKa = 10.92YY626 pKa = 10.81SDD628 pKa = 4.17LDD630 pKa = 3.53NTSDD634 pKa = 3.86YY635 pKa = 11.22KK636 pKa = 11.35AGDD639 pKa = 3.77YY640 pKa = 8.55VTADD644 pKa = 2.6IRR646 pKa = 11.84ANYY649 pKa = 9.6VIGDD653 pKa = 4.22FTLDD657 pKa = 3.25AYY659 pKa = 11.03VKK661 pKa = 10.58NLTNEE666 pKa = 4.79DD667 pKa = 2.66IVYY670 pKa = 9.06WDD672 pKa = 3.21NGAIEE677 pKa = 4.35RR678 pKa = 11.84VSVGQTRR685 pKa = 11.84TIGFNATFRR694 pKa = 11.84MM695 pKa = 4.24
MM1 pKa = 8.05PLTQFKK7 pKa = 10.49RR8 pKa = 11.84KK9 pKa = 9.24ALVVALVSQAGLTATNVAVANDD31 pKa = 3.58STEE34 pKa = 4.02IEE36 pKa = 4.11TDD38 pKa = 3.52TIVISGEE45 pKa = 3.86KK46 pKa = 9.38IEE48 pKa = 5.03RR49 pKa = 11.84SLKK52 pKa = 9.75EE53 pKa = 3.42ATTAVTVITEE63 pKa = 4.36DD64 pKa = 3.31QLQTTEE70 pKa = 4.01TQTINEE76 pKa = 4.38IAASTPNVINGGFGAVSIRR95 pKa = 11.84GVDD98 pKa = 3.48GTGAATGGYY107 pKa = 9.96AFYY110 pKa = 10.53SGARR114 pKa = 11.84ARR116 pKa = 11.84VSTIVDD122 pKa = 3.64GVSQSWSGYY131 pKa = 9.2NFTPSKK137 pKa = 10.44SWDD140 pKa = 3.41VKK142 pKa = 9.5QVEE145 pKa = 5.28VYY147 pKa = 10.44RR148 pKa = 11.84GPQSTTQGTNSIGGAMVVEE167 pKa = 4.96SNDD170 pKa = 3.03PTYY173 pKa = 10.57YY174 pKa = 10.2QEE176 pKa = 3.76AAIRR180 pKa = 11.84LGTEE184 pKa = 3.92VYY186 pKa = 10.79EE187 pKa = 4.69NGNTLSNLAVMASGPLIEE205 pKa = 5.86DD206 pKa = 3.21EE207 pKa = 4.39LAFRR211 pKa = 11.84LAVDD215 pKa = 4.33GSTGEE220 pKa = 4.12SYY222 pKa = 8.67ITYY225 pKa = 9.0EE226 pKa = 4.01QAATEE231 pKa = 4.59LDD233 pKa = 4.01DD234 pKa = 6.71SPDD237 pKa = 3.62LDD239 pKa = 3.46NTTNLNLRR247 pKa = 11.84SKK249 pKa = 10.49LLWEE253 pKa = 4.77PGFLPEE259 pKa = 5.01LSAKK263 pKa = 9.37ATVNYY268 pKa = 10.59SSFDD272 pKa = 3.29GSYY275 pKa = 11.06LNWANDD281 pKa = 3.19TDD283 pKa = 4.24EE284 pKa = 5.95DD285 pKa = 5.17YY286 pKa = 10.78STQTFTVSSADD297 pKa = 3.45RR298 pKa = 11.84DD299 pKa = 3.67YY300 pKa = 11.81TRR302 pKa = 11.84IQDD305 pKa = 3.72STVKK309 pKa = 10.51SVAGDD314 pKa = 3.46ADD316 pKa = 3.91YY317 pKa = 11.52QLTDD321 pKa = 3.38QISNSFQIVYY331 pKa = 10.81AEE333 pKa = 3.87TDD335 pKa = 3.56VEE337 pKa = 4.45FNEE340 pKa = 4.54YY341 pKa = 9.42TSSRR345 pKa = 11.84TVISDD350 pKa = 3.15VDD352 pKa = 3.58NVTVEE357 pKa = 3.51NRR359 pKa = 11.84LNFRR363 pKa = 11.84ASDD366 pKa = 3.7DD367 pKa = 3.65SATAFVGVYY376 pKa = 10.33LSNTEE381 pKa = 3.87TTQDD385 pKa = 3.12IFSVIDD391 pKa = 3.44VEE393 pKa = 5.21SDD395 pKa = 3.28VTTAAVFGEE404 pKa = 4.34ASYY407 pKa = 11.37QLLDD411 pKa = 4.07EE412 pKa = 5.12LSLIAGLRR420 pKa = 11.84YY421 pKa = 10.0EE422 pKa = 4.5NEE424 pKa = 4.01STDD427 pKa = 3.67RR428 pKa = 11.84MLDD431 pKa = 3.2HH432 pKa = 7.6ASNTNYY438 pKa = 10.33NFDD441 pKa = 3.62EE442 pKa = 4.52SVSEE446 pKa = 4.95DD447 pKa = 3.38VLLPKK452 pKa = 10.16IAAIYY457 pKa = 9.65EE458 pKa = 4.28LTDD461 pKa = 3.32NTVVSASVRR470 pKa = 11.84KK471 pKa = 9.91GYY473 pKa = 9.88NAGGGAVNWASDD485 pKa = 3.56TDD487 pKa = 3.93NIGYY491 pKa = 7.58YY492 pKa = 9.68TYY494 pKa = 11.1DD495 pKa = 3.53SEE497 pKa = 5.01TVWAYY502 pKa = 9.62EE503 pKa = 3.95AALKK507 pKa = 10.1TSIAGANVAVSAFINDD523 pKa = 3.51YY524 pKa = 11.33ADD526 pKa = 3.31YY527 pKa = 10.49QAFVSEE533 pKa = 4.57YY534 pKa = 10.77DD535 pKa = 3.35SSLDD539 pKa = 3.42DD540 pKa = 3.32TLQYY544 pKa = 10.44IEE546 pKa = 4.59NVEE549 pKa = 4.1DD550 pKa = 3.63ALTYY554 pKa = 10.36GVEE557 pKa = 4.1LEE559 pKa = 4.02GDD561 pKa = 3.09KK562 pKa = 10.67WLTDD566 pKa = 3.63STSLRR571 pKa = 11.84ASVGYY576 pKa = 7.78TQTEE580 pKa = 4.65VISDD584 pKa = 4.14DD585 pKa = 3.6DD586 pKa = 4.48TIDD589 pKa = 4.25GNEE592 pKa = 4.61LPNAPAFNTSVGVTQYY608 pKa = 10.78FSDD611 pKa = 4.01RR612 pKa = 11.84FSVGADD618 pKa = 2.97VVYY621 pKa = 10.84VGEE624 pKa = 4.51YY625 pKa = 10.92YY626 pKa = 10.81SDD628 pKa = 4.17LDD630 pKa = 3.53NTSDD634 pKa = 3.86YY635 pKa = 11.22KK636 pKa = 11.35AGDD639 pKa = 3.77YY640 pKa = 8.55VTADD644 pKa = 2.6IRR646 pKa = 11.84ANYY649 pKa = 9.6VIGDD653 pKa = 4.22FTLDD657 pKa = 3.25AYY659 pKa = 11.03VKK661 pKa = 10.58NLTNEE666 pKa = 4.79DD667 pKa = 2.66IVYY670 pKa = 9.06WDD672 pKa = 3.21NGAIEE677 pKa = 4.35RR678 pKa = 11.84VSVGQTRR685 pKa = 11.84TIGFNATFRR694 pKa = 11.84MM695 pKa = 4.24
Molecular weight: 75.61 kDa
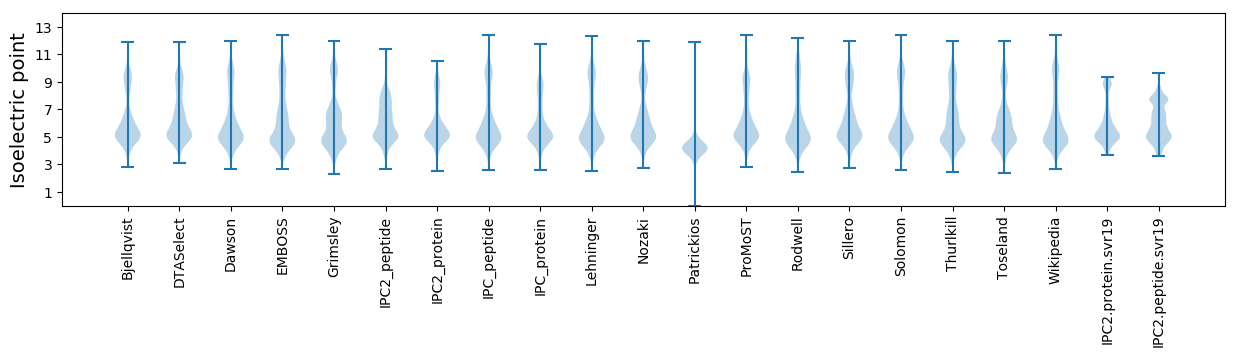
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R3HX17|A0A4R3HX17_9GAMM Transposase IS166 family protein OS=Reinekea marinisedimentorum OX=230495 GN=BCF53_12329 PE=4 SV=1
MM1 pKa = 7.19NRR3 pKa = 11.84ASNATIPGRR12 pKa = 11.84TWVKK16 pKa = 9.23RR17 pKa = 11.84TFYY20 pKa = 11.06LNRR23 pKa = 11.84SSLNDD28 pKa = 3.62MNHH31 pKa = 4.45VTQTLDD37 pKa = 3.08SALGIHH43 pKa = 6.4SVTFMPIQNSIEE55 pKa = 4.21VVFDD59 pKa = 3.58ARR61 pKa = 11.84LTCSQQIVAIFQQHH75 pKa = 6.37HH76 pKa = 6.06LPLRR80 pKa = 11.84AGLKK84 pKa = 10.16DD85 pKa = 3.26RR86 pKa = 11.84VRR88 pKa = 11.84FAWYY92 pKa = 10.09RR93 pKa = 11.84FIEE96 pKa = 4.18QNIKK100 pKa = 10.92DD101 pKa = 4.12NEE103 pKa = 4.26SSSRR107 pKa = 11.84WKK109 pKa = 10.25PLTPARR115 pKa = 4.25
MM1 pKa = 7.19NRR3 pKa = 11.84ASNATIPGRR12 pKa = 11.84TWVKK16 pKa = 9.23RR17 pKa = 11.84TFYY20 pKa = 11.06LNRR23 pKa = 11.84SSLNDD28 pKa = 3.62MNHH31 pKa = 4.45VTQTLDD37 pKa = 3.08SALGIHH43 pKa = 6.4SVTFMPIQNSIEE55 pKa = 4.21VVFDD59 pKa = 3.58ARR61 pKa = 11.84LTCSQQIVAIFQQHH75 pKa = 6.37HH76 pKa = 6.06LPLRR80 pKa = 11.84AGLKK84 pKa = 10.16DD85 pKa = 3.26RR86 pKa = 11.84VRR88 pKa = 11.84FAWYY92 pKa = 10.09RR93 pKa = 11.84FIEE96 pKa = 4.18QNIKK100 pKa = 10.92DD101 pKa = 4.12NEE103 pKa = 4.26SSSRR107 pKa = 11.84WKK109 pKa = 10.25PLTPARR115 pKa = 4.25
Molecular weight: 13.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1253563 |
29 |
2328 |
329.9 |
36.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.194 ± 0.04 | 1.031 ± 0.015 |
5.659 ± 0.046 | 6.583 ± 0.038 |
4.102 ± 0.027 | 6.89 ± 0.033 |
2.122 ± 0.019 | 6.105 ± 0.028 |
4.757 ± 0.032 | 10.376 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.476 ± 0.018 | 4.108 ± 0.028 |
3.971 ± 0.027 | 4.386 ± 0.033 |
4.699 ± 0.032 | 6.795 ± 0.036 |
5.345 ± 0.029 | 6.979 ± 0.031 |
1.334 ± 0.016 | 3.088 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
