
Vibrio phage Marilyn
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
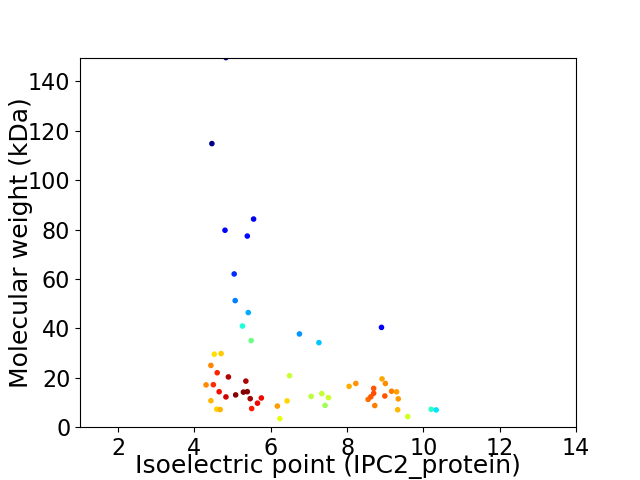
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M9YZD6|A0A6M9YZD6_9CAUD Uncharacterized protein OS=Vibrio phage Marilyn OX=2736287 GN=MARILYN_24 PE=4 SV=1
MM1 pKa = 7.41SKK3 pKa = 8.1TWIAKK8 pKa = 9.13CVSFKK13 pKa = 10.88GKK15 pKa = 9.67HH16 pKa = 4.75PQGEE20 pKa = 4.34VTHH23 pKa = 6.14RR24 pKa = 11.84VVVKK28 pKa = 10.46AQYY31 pKa = 10.54KK32 pKa = 9.85KK33 pKa = 10.92DD34 pKa = 3.82GIEE37 pKa = 3.92AAQEE41 pKa = 3.82EE42 pKa = 5.0LKK44 pKa = 11.22LFDD47 pKa = 4.31VEE49 pKa = 5.25HH50 pKa = 6.6HH51 pKa = 7.13DD52 pKa = 4.01KK53 pKa = 11.07YY54 pKa = 11.13KK55 pKa = 10.76KK56 pKa = 10.08PEE58 pKa = 3.94LTEE61 pKa = 4.1VMDD64 pKa = 3.71EE65 pKa = 4.4SEE67 pKa = 4.37AEE69 pKa = 4.1RR70 pKa = 11.84IYY72 pKa = 10.51IEE74 pKa = 4.08YY75 pKa = 9.01MAVRR79 pKa = 11.84EE80 pKa = 3.83QLEE83 pKa = 4.12QIDD86 pKa = 4.29LCDD89 pKa = 4.95DD90 pKa = 3.47EE91 pKa = 6.59SNFNDD96 pKa = 3.52VEE98 pKa = 4.2QEE100 pKa = 3.84KK101 pKa = 10.05TDD103 pKa = 3.5AYY105 pKa = 10.63MEE107 pKa = 4.26ATQTLLARR115 pKa = 11.84YY116 pKa = 9.29DD117 pKa = 3.44IIDD120 pKa = 3.74RR121 pKa = 11.84VTVNHH126 pKa = 6.31IMEE129 pKa = 4.53IRR131 pKa = 11.84NKK133 pKa = 10.0LEE135 pKa = 3.73EE136 pKa = 4.23VIKK139 pKa = 10.67EE140 pKa = 4.07NEE142 pKa = 4.06EE143 pKa = 3.91CDD145 pKa = 4.52PIATLEE151 pKa = 4.52NIASYY156 pKa = 10.05NWKK159 pKa = 9.44QRR161 pKa = 11.84FPMGTDD167 pKa = 2.7EE168 pKa = 6.53KK169 pKa = 10.43MLQVLLNRR177 pKa = 11.84RR178 pKa = 11.84EE179 pKa = 3.67MRR181 pKa = 11.84AALDD185 pKa = 3.17GMYY188 pKa = 9.74IEE190 pKa = 4.81KK191 pKa = 8.09TTTEE195 pKa = 4.25EE196 pKa = 3.95PSEE199 pKa = 3.98EE200 pKa = 4.01LVDD203 pKa = 3.83SGEE206 pKa = 4.11KK207 pKa = 8.89FTAEE211 pKa = 3.78KK212 pKa = 9.74ATIAEE217 pKa = 4.16YY218 pKa = 9.42PIIEE222 pKa = 4.78PYY224 pKa = 9.67SVRR227 pKa = 11.84IAVAKK232 pKa = 9.61QGEE235 pKa = 4.72SFVHH239 pKa = 6.42SMAVVNRR246 pKa = 11.84EE247 pKa = 3.55KK248 pKa = 10.97GGNVYY253 pKa = 10.42GQLDD257 pKa = 3.76VFNPKK262 pKa = 10.06EE263 pKa = 3.9FTNVEE268 pKa = 4.06NAVVYY273 pKa = 10.23ALAMSIEE280 pKa = 4.35AVNKK284 pKa = 8.72YY285 pKa = 9.96LPHH288 pKa = 6.77FLEE291 pKa = 4.29EE292 pKa = 4.23RR293 pKa = 11.84QVNTCVNILSSYY305 pKa = 9.87AQDD308 pKa = 3.36AEE310 pKa = 4.65GNFIEE315 pKa = 6.26RR316 pKa = 11.84YY317 pKa = 9.21MGDD320 pKa = 3.51HH321 pKa = 7.26DD322 pKa = 5.27LSNATFMAVSQPKK335 pKa = 9.96EE336 pKa = 3.74EE337 pKa = 4.59SKK339 pKa = 10.74QSSGKK344 pKa = 10.03EE345 pKa = 3.83VTSDD349 pKa = 3.27VARR352 pKa = 11.84ITAAAVAHH360 pKa = 6.34PSIAPHH366 pKa = 6.2MEE368 pKa = 3.55TDD370 pKa = 3.22NRR372 pKa = 11.84AKK374 pKa = 10.13EE375 pKa = 3.99VYY377 pKa = 10.13YY378 pKa = 10.0QIQEE382 pKa = 4.15NFTALHH388 pKa = 5.83NKK390 pKa = 10.02YY391 pKa = 10.26GFLPAAQMVEE401 pKa = 4.74HH402 pKa = 7.47IISMSDD408 pKa = 3.13VSDD411 pKa = 5.28LITIEE416 pKa = 4.4GCDD419 pKa = 3.92EE420 pKa = 4.12LASHH424 pKa = 5.93YY425 pKa = 10.99VEE427 pKa = 4.24NTTTQTMQAITEE439 pKa = 4.2SSEE442 pKa = 3.82EE443 pKa = 3.97EE444 pKa = 3.99NDD446 pKa = 3.3VKK448 pKa = 10.84FYY450 pKa = 11.13AGQVWRR456 pKa = 11.84CYY458 pKa = 10.01IEE460 pKa = 5.41EE461 pKa = 4.02ILAQFKK467 pKa = 8.98TEE469 pKa = 3.99KK470 pKa = 10.3QISSEE475 pKa = 4.76DD476 pKa = 2.95IDD478 pKa = 4.22AAAKK482 pKa = 10.35NISRR486 pKa = 11.84LFNDD490 pKa = 3.32WKK492 pKa = 11.1LSLFKK497 pKa = 10.95EE498 pKa = 4.05KK499 pKa = 10.7GKK501 pKa = 8.92YY502 pKa = 8.72TIDD505 pKa = 3.27ADD507 pKa = 3.78MFKK510 pKa = 11.19DD511 pKa = 3.99EE512 pKa = 5.43IYY514 pKa = 10.92LIFNNSEE521 pKa = 3.88YY522 pKa = 10.84QKK524 pKa = 11.35ALEE527 pKa = 4.05LLKK530 pKa = 10.72DD531 pKa = 3.67YY532 pKa = 10.81FGSKK536 pKa = 10.01DD537 pKa = 3.28QLVNVLACGFDD548 pKa = 4.02DD549 pKa = 5.36YY550 pKa = 12.11VNEE553 pKa = 4.38LNPKK557 pKa = 9.47VKK559 pKa = 10.59EE560 pKa = 3.94EE561 pKa = 3.91LVNLVNRR568 pKa = 11.84LAVDD572 pKa = 3.71SSLGCTHH579 pKa = 6.64TSEE582 pKa = 4.43VVKK585 pKa = 10.86ALIYY589 pKa = 10.65SEE591 pKa = 5.35LSAEE595 pKa = 4.39SVPDD599 pKa = 3.6MVILKK604 pKa = 9.75PNLKK608 pKa = 9.22MDD610 pKa = 4.04FWNKK614 pKa = 9.87LNHH617 pKa = 7.13DD618 pKa = 3.87YY619 pKa = 11.12CSNQRR624 pKa = 11.84QRR626 pKa = 11.84QEE628 pKa = 4.06EE629 pKa = 4.55TVGDD633 pKa = 3.7YY634 pKa = 11.59VMAYY638 pKa = 10.02GSSDD642 pKa = 3.56EE643 pKa = 4.76LRR645 pKa = 11.84AKK647 pKa = 10.71ANEE650 pKa = 3.61IDD652 pKa = 4.08AKK654 pKa = 10.35AKK656 pKa = 9.97EE657 pKa = 4.5YY658 pKa = 10.54INEE661 pKa = 3.94VCEE664 pKa = 4.28PLAQALNEE672 pKa = 4.14KK673 pKa = 9.21VQEE676 pKa = 4.24VNAKK680 pKa = 10.11AEE682 pKa = 4.23EE683 pKa = 4.39YY684 pKa = 10.61EE685 pKa = 4.13NEE687 pKa = 3.87ACVRR691 pKa = 11.84GDD693 pKa = 3.16GVTPEE698 pKa = 4.35SEE700 pKa = 4.44RR701 pKa = 11.84FTIDD705 pKa = 3.4APKK708 pKa = 9.42TLPDD712 pKa = 3.67APEE715 pKa = 4.02LRR717 pKa = 11.84EE718 pKa = 3.85GATIGNSVEE727 pKa = 4.17ITDD730 pKa = 4.11EE731 pKa = 4.36EE732 pKa = 4.89YY733 pKa = 10.89SALLNPEE740 pKa = 4.83DD741 pKa = 3.87VPEE744 pKa = 4.05PQILPDD750 pKa = 3.89DD751 pKa = 3.79MEE753 pKa = 4.34VRR755 pKa = 11.84MEE757 pKa = 3.81EE758 pKa = 3.95TDD760 pKa = 2.97ASKK763 pKa = 11.39YY764 pKa = 10.8YY765 pKa = 9.49GTALLNGDD773 pKa = 3.21IDD775 pKa = 4.03SPYY778 pKa = 10.04EE779 pKa = 3.6LAIAYY784 pKa = 7.5MMKK787 pKa = 10.62DD788 pKa = 3.48LGAEE792 pKa = 4.14NYY794 pKa = 10.01HH795 pKa = 6.8DD796 pKa = 4.61AGWYY800 pKa = 9.28QYY802 pKa = 11.32AAAKK806 pKa = 10.24LRR808 pKa = 11.84GAAEE812 pKa = 4.08RR813 pKa = 11.84LKK815 pKa = 10.87CDD817 pKa = 4.45LPALMTGLTVEE828 pKa = 4.05KK829 pKa = 10.32LVRR832 pKa = 11.84EE833 pKa = 4.07KK834 pKa = 10.6SWANVEE840 pKa = 4.13EE841 pKa = 4.57LEE843 pKa = 4.54QALEE847 pKa = 3.97SHH849 pKa = 7.18EE850 pKa = 4.32ILDD853 pKa = 3.86TALYY857 pKa = 8.43EE858 pKa = 4.49HH859 pKa = 6.94GVFDD863 pKa = 4.04YY864 pKa = 11.32DD865 pKa = 3.85EE866 pKa = 4.51EE867 pKa = 4.77VLFGTDD873 pKa = 3.32HH874 pKa = 5.53QQEE877 pKa = 4.3APADD881 pKa = 3.9EE882 pKa = 4.84PEE884 pKa = 4.35EE885 pKa = 3.96ARR887 pKa = 11.84QDD889 pKa = 3.58IEE891 pKa = 4.73PDD893 pKa = 2.95NDD895 pKa = 4.1DD896 pKa = 4.48PVTSADD902 pKa = 3.82NGDD905 pKa = 4.25SVATDD910 pKa = 3.27NEE912 pKa = 4.23LSVRR916 pKa = 11.84EE917 pKa = 3.93LFKK920 pKa = 11.06RR921 pKa = 11.84QMAAANQKK929 pKa = 10.32LGLVGFAWGYY939 pKa = 6.51EE940 pKa = 4.38VKK942 pKa = 10.69RR943 pKa = 11.84QWTEE947 pKa = 3.53DD948 pKa = 3.5KK949 pKa = 10.95KK950 pKa = 11.04DD951 pKa = 3.26GRR953 pKa = 11.84TDD955 pKa = 3.11CHH957 pKa = 7.99VEE959 pKa = 3.59VGIWFKK965 pKa = 11.36DD966 pKa = 3.33KK967 pKa = 11.27NGTKK971 pKa = 10.43GEE973 pKa = 4.51PISFYY978 pKa = 11.04GVSNANFGNYY988 pKa = 9.18VNDD991 pKa = 3.32AMMLAVVNGLEE1002 pKa = 3.83ITLISLGVEE1011 pKa = 4.23VEE1013 pKa = 4.37GG1014 pKa = 5.11
MM1 pKa = 7.41SKK3 pKa = 8.1TWIAKK8 pKa = 9.13CVSFKK13 pKa = 10.88GKK15 pKa = 9.67HH16 pKa = 4.75PQGEE20 pKa = 4.34VTHH23 pKa = 6.14RR24 pKa = 11.84VVVKK28 pKa = 10.46AQYY31 pKa = 10.54KK32 pKa = 9.85KK33 pKa = 10.92DD34 pKa = 3.82GIEE37 pKa = 3.92AAQEE41 pKa = 3.82EE42 pKa = 5.0LKK44 pKa = 11.22LFDD47 pKa = 4.31VEE49 pKa = 5.25HH50 pKa = 6.6HH51 pKa = 7.13DD52 pKa = 4.01KK53 pKa = 11.07YY54 pKa = 11.13KK55 pKa = 10.76KK56 pKa = 10.08PEE58 pKa = 3.94LTEE61 pKa = 4.1VMDD64 pKa = 3.71EE65 pKa = 4.4SEE67 pKa = 4.37AEE69 pKa = 4.1RR70 pKa = 11.84IYY72 pKa = 10.51IEE74 pKa = 4.08YY75 pKa = 9.01MAVRR79 pKa = 11.84EE80 pKa = 3.83QLEE83 pKa = 4.12QIDD86 pKa = 4.29LCDD89 pKa = 4.95DD90 pKa = 3.47EE91 pKa = 6.59SNFNDD96 pKa = 3.52VEE98 pKa = 4.2QEE100 pKa = 3.84KK101 pKa = 10.05TDD103 pKa = 3.5AYY105 pKa = 10.63MEE107 pKa = 4.26ATQTLLARR115 pKa = 11.84YY116 pKa = 9.29DD117 pKa = 3.44IIDD120 pKa = 3.74RR121 pKa = 11.84VTVNHH126 pKa = 6.31IMEE129 pKa = 4.53IRR131 pKa = 11.84NKK133 pKa = 10.0LEE135 pKa = 3.73EE136 pKa = 4.23VIKK139 pKa = 10.67EE140 pKa = 4.07NEE142 pKa = 4.06EE143 pKa = 3.91CDD145 pKa = 4.52PIATLEE151 pKa = 4.52NIASYY156 pKa = 10.05NWKK159 pKa = 9.44QRR161 pKa = 11.84FPMGTDD167 pKa = 2.7EE168 pKa = 6.53KK169 pKa = 10.43MLQVLLNRR177 pKa = 11.84RR178 pKa = 11.84EE179 pKa = 3.67MRR181 pKa = 11.84AALDD185 pKa = 3.17GMYY188 pKa = 9.74IEE190 pKa = 4.81KK191 pKa = 8.09TTTEE195 pKa = 4.25EE196 pKa = 3.95PSEE199 pKa = 3.98EE200 pKa = 4.01LVDD203 pKa = 3.83SGEE206 pKa = 4.11KK207 pKa = 8.89FTAEE211 pKa = 3.78KK212 pKa = 9.74ATIAEE217 pKa = 4.16YY218 pKa = 9.42PIIEE222 pKa = 4.78PYY224 pKa = 9.67SVRR227 pKa = 11.84IAVAKK232 pKa = 9.61QGEE235 pKa = 4.72SFVHH239 pKa = 6.42SMAVVNRR246 pKa = 11.84EE247 pKa = 3.55KK248 pKa = 10.97GGNVYY253 pKa = 10.42GQLDD257 pKa = 3.76VFNPKK262 pKa = 10.06EE263 pKa = 3.9FTNVEE268 pKa = 4.06NAVVYY273 pKa = 10.23ALAMSIEE280 pKa = 4.35AVNKK284 pKa = 8.72YY285 pKa = 9.96LPHH288 pKa = 6.77FLEE291 pKa = 4.29EE292 pKa = 4.23RR293 pKa = 11.84QVNTCVNILSSYY305 pKa = 9.87AQDD308 pKa = 3.36AEE310 pKa = 4.65GNFIEE315 pKa = 6.26RR316 pKa = 11.84YY317 pKa = 9.21MGDD320 pKa = 3.51HH321 pKa = 7.26DD322 pKa = 5.27LSNATFMAVSQPKK335 pKa = 9.96EE336 pKa = 3.74EE337 pKa = 4.59SKK339 pKa = 10.74QSSGKK344 pKa = 10.03EE345 pKa = 3.83VTSDD349 pKa = 3.27VARR352 pKa = 11.84ITAAAVAHH360 pKa = 6.34PSIAPHH366 pKa = 6.2MEE368 pKa = 3.55TDD370 pKa = 3.22NRR372 pKa = 11.84AKK374 pKa = 10.13EE375 pKa = 3.99VYY377 pKa = 10.13YY378 pKa = 10.0QIQEE382 pKa = 4.15NFTALHH388 pKa = 5.83NKK390 pKa = 10.02YY391 pKa = 10.26GFLPAAQMVEE401 pKa = 4.74HH402 pKa = 7.47IISMSDD408 pKa = 3.13VSDD411 pKa = 5.28LITIEE416 pKa = 4.4GCDD419 pKa = 3.92EE420 pKa = 4.12LASHH424 pKa = 5.93YY425 pKa = 10.99VEE427 pKa = 4.24NTTTQTMQAITEE439 pKa = 4.2SSEE442 pKa = 3.82EE443 pKa = 3.97EE444 pKa = 3.99NDD446 pKa = 3.3VKK448 pKa = 10.84FYY450 pKa = 11.13AGQVWRR456 pKa = 11.84CYY458 pKa = 10.01IEE460 pKa = 5.41EE461 pKa = 4.02ILAQFKK467 pKa = 8.98TEE469 pKa = 3.99KK470 pKa = 10.3QISSEE475 pKa = 4.76DD476 pKa = 2.95IDD478 pKa = 4.22AAAKK482 pKa = 10.35NISRR486 pKa = 11.84LFNDD490 pKa = 3.32WKK492 pKa = 11.1LSLFKK497 pKa = 10.95EE498 pKa = 4.05KK499 pKa = 10.7GKK501 pKa = 8.92YY502 pKa = 8.72TIDD505 pKa = 3.27ADD507 pKa = 3.78MFKK510 pKa = 11.19DD511 pKa = 3.99EE512 pKa = 5.43IYY514 pKa = 10.92LIFNNSEE521 pKa = 3.88YY522 pKa = 10.84QKK524 pKa = 11.35ALEE527 pKa = 4.05LLKK530 pKa = 10.72DD531 pKa = 3.67YY532 pKa = 10.81FGSKK536 pKa = 10.01DD537 pKa = 3.28QLVNVLACGFDD548 pKa = 4.02DD549 pKa = 5.36YY550 pKa = 12.11VNEE553 pKa = 4.38LNPKK557 pKa = 9.47VKK559 pKa = 10.59EE560 pKa = 3.94EE561 pKa = 3.91LVNLVNRR568 pKa = 11.84LAVDD572 pKa = 3.71SSLGCTHH579 pKa = 6.64TSEE582 pKa = 4.43VVKK585 pKa = 10.86ALIYY589 pKa = 10.65SEE591 pKa = 5.35LSAEE595 pKa = 4.39SVPDD599 pKa = 3.6MVILKK604 pKa = 9.75PNLKK608 pKa = 9.22MDD610 pKa = 4.04FWNKK614 pKa = 9.87LNHH617 pKa = 7.13DD618 pKa = 3.87YY619 pKa = 11.12CSNQRR624 pKa = 11.84QRR626 pKa = 11.84QEE628 pKa = 4.06EE629 pKa = 4.55TVGDD633 pKa = 3.7YY634 pKa = 11.59VMAYY638 pKa = 10.02GSSDD642 pKa = 3.56EE643 pKa = 4.76LRR645 pKa = 11.84AKK647 pKa = 10.71ANEE650 pKa = 3.61IDD652 pKa = 4.08AKK654 pKa = 10.35AKK656 pKa = 9.97EE657 pKa = 4.5YY658 pKa = 10.54INEE661 pKa = 3.94VCEE664 pKa = 4.28PLAQALNEE672 pKa = 4.14KK673 pKa = 9.21VQEE676 pKa = 4.24VNAKK680 pKa = 10.11AEE682 pKa = 4.23EE683 pKa = 4.39YY684 pKa = 10.61EE685 pKa = 4.13NEE687 pKa = 3.87ACVRR691 pKa = 11.84GDD693 pKa = 3.16GVTPEE698 pKa = 4.35SEE700 pKa = 4.44RR701 pKa = 11.84FTIDD705 pKa = 3.4APKK708 pKa = 9.42TLPDD712 pKa = 3.67APEE715 pKa = 4.02LRR717 pKa = 11.84EE718 pKa = 3.85GATIGNSVEE727 pKa = 4.17ITDD730 pKa = 4.11EE731 pKa = 4.36EE732 pKa = 4.89YY733 pKa = 10.89SALLNPEE740 pKa = 4.83DD741 pKa = 3.87VPEE744 pKa = 4.05PQILPDD750 pKa = 3.89DD751 pKa = 3.79MEE753 pKa = 4.34VRR755 pKa = 11.84MEE757 pKa = 3.81EE758 pKa = 3.95TDD760 pKa = 2.97ASKK763 pKa = 11.39YY764 pKa = 10.8YY765 pKa = 9.49GTALLNGDD773 pKa = 3.21IDD775 pKa = 4.03SPYY778 pKa = 10.04EE779 pKa = 3.6LAIAYY784 pKa = 7.5MMKK787 pKa = 10.62DD788 pKa = 3.48LGAEE792 pKa = 4.14NYY794 pKa = 10.01HH795 pKa = 6.8DD796 pKa = 4.61AGWYY800 pKa = 9.28QYY802 pKa = 11.32AAAKK806 pKa = 10.24LRR808 pKa = 11.84GAAEE812 pKa = 4.08RR813 pKa = 11.84LKK815 pKa = 10.87CDD817 pKa = 4.45LPALMTGLTVEE828 pKa = 4.05KK829 pKa = 10.32LVRR832 pKa = 11.84EE833 pKa = 4.07KK834 pKa = 10.6SWANVEE840 pKa = 4.13EE841 pKa = 4.57LEE843 pKa = 4.54QALEE847 pKa = 3.97SHH849 pKa = 7.18EE850 pKa = 4.32ILDD853 pKa = 3.86TALYY857 pKa = 8.43EE858 pKa = 4.49HH859 pKa = 6.94GVFDD863 pKa = 4.04YY864 pKa = 11.32DD865 pKa = 3.85EE866 pKa = 4.51EE867 pKa = 4.77VLFGTDD873 pKa = 3.32HH874 pKa = 5.53QQEE877 pKa = 4.3APADD881 pKa = 3.9EE882 pKa = 4.84PEE884 pKa = 4.35EE885 pKa = 3.96ARR887 pKa = 11.84QDD889 pKa = 3.58IEE891 pKa = 4.73PDD893 pKa = 2.95NDD895 pKa = 4.1DD896 pKa = 4.48PVTSADD902 pKa = 3.82NGDD905 pKa = 4.25SVATDD910 pKa = 3.27NEE912 pKa = 4.23LSVRR916 pKa = 11.84EE917 pKa = 3.93LFKK920 pKa = 11.06RR921 pKa = 11.84QMAAANQKK929 pKa = 10.32LGLVGFAWGYY939 pKa = 6.51EE940 pKa = 4.38VKK942 pKa = 10.69RR943 pKa = 11.84QWTEE947 pKa = 3.53DD948 pKa = 3.5KK949 pKa = 10.95KK950 pKa = 11.04DD951 pKa = 3.26GRR953 pKa = 11.84TDD955 pKa = 3.11CHH957 pKa = 7.99VEE959 pKa = 3.59VGIWFKK965 pKa = 11.36DD966 pKa = 3.33KK967 pKa = 11.27NGTKK971 pKa = 10.43GEE973 pKa = 4.51PISFYY978 pKa = 11.04GVSNANFGNYY988 pKa = 9.18VNDD991 pKa = 3.32AMMLAVVNGLEE1002 pKa = 3.83ITLISLGVEE1011 pKa = 4.23VEE1013 pKa = 4.37GG1014 pKa = 5.11
Molecular weight: 114.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M9YZF3|A0A6M9YZF3_9CAUD Uncharacterized protein OS=Vibrio phage Marilyn OX=2736287 GN=MARILYN_54 PE=4 SV=1
MM1 pKa = 7.17FLIGAGNAVLRR12 pKa = 11.84KK13 pKa = 10.09NIFTTSVAKK22 pKa = 10.44LHH24 pKa = 5.86FKK26 pKa = 10.5KK27 pKa = 10.76VLSNFGGQGAPVGNHH42 pKa = 4.87GRR44 pKa = 11.84LKK46 pKa = 10.37RR47 pKa = 11.84NYY49 pKa = 8.36EE50 pKa = 4.13FPHH53 pKa = 6.56PAHH56 pKa = 6.72HH57 pKa = 7.17PFAIPHH63 pKa = 5.77IWW65 pKa = 3.02
MM1 pKa = 7.17FLIGAGNAVLRR12 pKa = 11.84KK13 pKa = 10.09NIFTTSVAKK22 pKa = 10.44LHH24 pKa = 5.86FKK26 pKa = 10.5KK27 pKa = 10.76VLSNFGGQGAPVGNHH42 pKa = 4.87GRR44 pKa = 11.84LKK46 pKa = 10.37RR47 pKa = 11.84NYY49 pKa = 8.36EE50 pKa = 4.13FPHH53 pKa = 6.56PAHH56 pKa = 6.72HH57 pKa = 7.17PFAIPHH63 pKa = 5.77IWW65 pKa = 3.02
Molecular weight: 7.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13179 |
31 |
1375 |
227.2 |
25.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.992 ± 0.453 | 0.865 ± 0.122 |
6.237 ± 0.254 | 7.398 ± 0.458 |
3.65 ± 0.19 | 6.844 ± 0.372 |
1.874 ± 0.194 | 5.911 ± 0.246 |
6.806 ± 0.276 | 7.542 ± 0.384 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.58 ± 0.198 | 5.145 ± 0.214 |
3.703 ± 0.211 | 3.991 ± 0.202 |
4.796 ± 0.312 | 6.571 ± 0.339 |
6.047 ± 0.263 | 6.252 ± 0.274 |
1.404 ± 0.115 | 3.392 ± 0.248 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
