
Isoptericola dokdonensis DS-3
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Isoptericola; Isoptericola dokdonensis
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
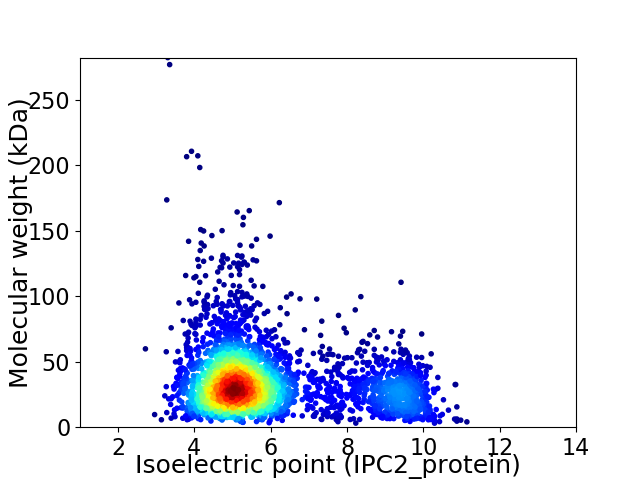
Virtual 2D-PAGE plot for 3403 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161III5|A0A161III5_9MICO Uncharacterized protein OS=Isoptericola dokdonensis DS-3 OX=1300344 GN=I598_2183 PE=4 SV=1
MM1 pKa = 6.89PTLRR5 pKa = 11.84RR6 pKa = 11.84ALAATAALALPAVALTACSTDD27 pKa = 3.47GEE29 pKa = 4.97GTGSDD34 pKa = 3.96DD35 pKa = 3.81GTLQVLASFYY45 pKa = 9.4PLQYY49 pKa = 10.07VAEE52 pKa = 4.2QVGGDD57 pKa = 3.96LVSVDD62 pKa = 3.61NLTPPSAEE70 pKa = 3.84PHH72 pKa = 6.29DD73 pKa = 5.34LEE75 pKa = 5.13LAPAQVRR82 pKa = 11.84AVGDD86 pKa = 3.67ADD88 pKa = 3.76LVVYY92 pKa = 10.08QSGFQAAVDD101 pKa = 3.78EE102 pKa = 4.89AVDD105 pKa = 3.5ARR107 pKa = 11.84QPEE110 pKa = 4.55HH111 pKa = 7.28VVDD114 pKa = 3.83AADD117 pKa = 3.72VVEE120 pKa = 5.44LEE122 pKa = 4.32ASPGTVEE129 pKa = 5.45HH130 pKa = 6.91MDD132 pKa = 4.24DD133 pKa = 3.34EE134 pKa = 5.87HH135 pKa = 8.17EE136 pKa = 4.74GEE138 pKa = 4.3TAEE141 pKa = 4.23EE142 pKa = 4.23HH143 pKa = 6.97AEE145 pKa = 4.1HH146 pKa = 7.17ADD148 pKa = 3.55EE149 pKa = 5.47EE150 pKa = 4.8DD151 pKa = 3.86GHH153 pKa = 7.6DD154 pKa = 4.29HH155 pKa = 7.02GALDD159 pKa = 3.54PHH161 pKa = 6.79FWLDD165 pKa = 3.49PTLLEE170 pKa = 4.26PVADD174 pKa = 3.89AVADD178 pKa = 3.92EE179 pKa = 4.98LSAVDD184 pKa = 4.16PGNAATYY191 pKa = 9.59AANAEE196 pKa = 4.29ALTATLDD203 pKa = 4.19DD204 pKa = 4.27LDD206 pKa = 4.22ARR208 pKa = 11.84YY209 pKa = 7.96ATALEE214 pKa = 4.42PCAGQTLVTSHH225 pKa = 6.1TAFGYY230 pKa = 9.82LAEE233 pKa = 5.31RR234 pKa = 11.84YY235 pKa = 9.78DD236 pKa = 3.81LVQIGIAAIDD246 pKa = 4.1PEE248 pKa = 4.9AEE250 pKa = 4.02PSPARR255 pKa = 11.84LRR257 pKa = 11.84EE258 pKa = 3.97IGDD261 pKa = 3.51VVEE264 pKa = 5.01ANDD267 pKa = 3.32VQTIFTEE274 pKa = 4.54TLTSPKK280 pKa = 10.08VAEE283 pKa = 4.42TLASDD288 pKa = 4.34LGVATAVLDD297 pKa = 4.07PLEE300 pKa = 4.46GLSTEE305 pKa = 4.37AADD308 pKa = 4.51AGADD312 pKa = 3.68YY313 pKa = 10.81VAVMDD318 pKa = 5.58DD319 pKa = 3.99NLDD322 pKa = 3.75SLVTGLGCDD331 pKa = 3.22AA332 pKa = 5.43
MM1 pKa = 6.89PTLRR5 pKa = 11.84RR6 pKa = 11.84ALAATAALALPAVALTACSTDD27 pKa = 3.47GEE29 pKa = 4.97GTGSDD34 pKa = 3.96DD35 pKa = 3.81GTLQVLASFYY45 pKa = 9.4PLQYY49 pKa = 10.07VAEE52 pKa = 4.2QVGGDD57 pKa = 3.96LVSVDD62 pKa = 3.61NLTPPSAEE70 pKa = 3.84PHH72 pKa = 6.29DD73 pKa = 5.34LEE75 pKa = 5.13LAPAQVRR82 pKa = 11.84AVGDD86 pKa = 3.67ADD88 pKa = 3.76LVVYY92 pKa = 10.08QSGFQAAVDD101 pKa = 3.78EE102 pKa = 4.89AVDD105 pKa = 3.5ARR107 pKa = 11.84QPEE110 pKa = 4.55HH111 pKa = 7.28VVDD114 pKa = 3.83AADD117 pKa = 3.72VVEE120 pKa = 5.44LEE122 pKa = 4.32ASPGTVEE129 pKa = 5.45HH130 pKa = 6.91MDD132 pKa = 4.24DD133 pKa = 3.34EE134 pKa = 5.87HH135 pKa = 8.17EE136 pKa = 4.74GEE138 pKa = 4.3TAEE141 pKa = 4.23EE142 pKa = 4.23HH143 pKa = 6.97AEE145 pKa = 4.1HH146 pKa = 7.17ADD148 pKa = 3.55EE149 pKa = 5.47EE150 pKa = 4.8DD151 pKa = 3.86GHH153 pKa = 7.6DD154 pKa = 4.29HH155 pKa = 7.02GALDD159 pKa = 3.54PHH161 pKa = 6.79FWLDD165 pKa = 3.49PTLLEE170 pKa = 4.26PVADD174 pKa = 3.89AVADD178 pKa = 3.92EE179 pKa = 4.98LSAVDD184 pKa = 4.16PGNAATYY191 pKa = 9.59AANAEE196 pKa = 4.29ALTATLDD203 pKa = 4.19DD204 pKa = 4.27LDD206 pKa = 4.22ARR208 pKa = 11.84YY209 pKa = 7.96ATALEE214 pKa = 4.42PCAGQTLVTSHH225 pKa = 6.1TAFGYY230 pKa = 9.82LAEE233 pKa = 5.31RR234 pKa = 11.84YY235 pKa = 9.78DD236 pKa = 3.81LVQIGIAAIDD246 pKa = 4.1PEE248 pKa = 4.9AEE250 pKa = 4.02PSPARR255 pKa = 11.84LRR257 pKa = 11.84EE258 pKa = 3.97IGDD261 pKa = 3.51VVEE264 pKa = 5.01ANDD267 pKa = 3.32VQTIFTEE274 pKa = 4.54TLTSPKK280 pKa = 10.08VAEE283 pKa = 4.42TLASDD288 pKa = 4.34LGVATAVLDD297 pKa = 4.07PLEE300 pKa = 4.46GLSTEE305 pKa = 4.37AADD308 pKa = 4.51AGADD312 pKa = 3.68YY313 pKa = 10.81VAVMDD318 pKa = 5.58DD319 pKa = 3.99NLDD322 pKa = 3.75SLVTGLGCDD331 pKa = 3.22AA332 pKa = 5.43
Molecular weight: 34.43 kDa
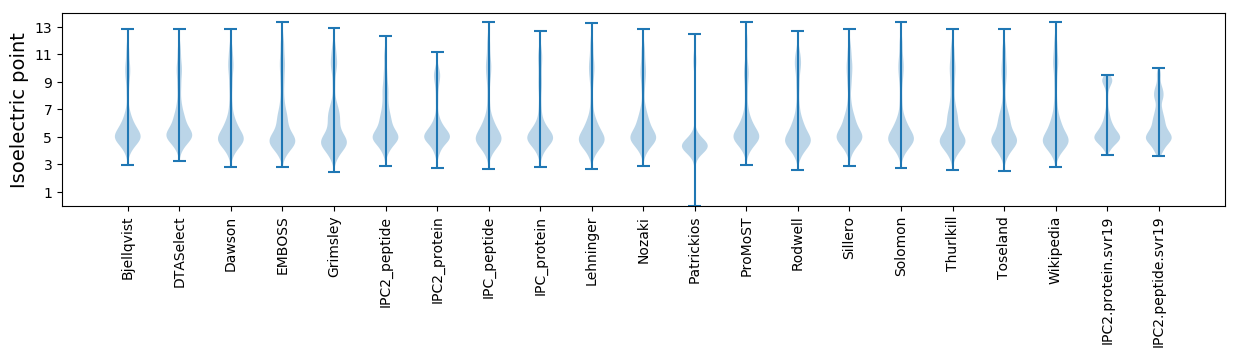
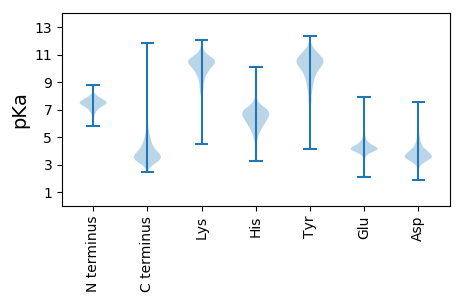
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161II59|A0A161II59_9MICO HTH cro/C1-type domain-containing protein OS=Isoptericola dokdonensis DS-3 OX=1300344 GN=I598_0107 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1153230 |
29 |
2802 |
338.9 |
35.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.449 ± 0.067 | 0.542 ± 0.009 |
6.947 ± 0.039 | 5.265 ± 0.04 |
2.528 ± 0.022 | 9.5 ± 0.04 |
2.137 ± 0.022 | 2.633 ± 0.031 |
1.427 ± 0.028 | 10.124 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.537 ± 0.016 | 1.466 ± 0.025 |
5.948 ± 0.039 | 2.515 ± 0.02 |
7.776 ± 0.061 | 4.754 ± 0.035 |
6.554 ± 0.044 | 10.525 ± 0.053 |
1.576 ± 0.019 | 1.797 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
