
Lake Sarah-associated circular virus-19
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.12
Get precalculated fractions of proteins
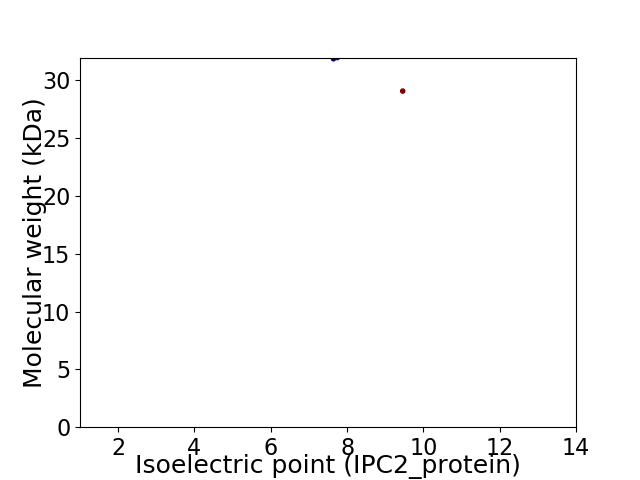
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G9N6|A0A126G9N6_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-19 OX=1685745 PE=4 SV=1
MM1 pKa = 8.25DD2 pKa = 6.29DD3 pKa = 3.89YY4 pKa = 11.69LLKK7 pKa = 10.68FCEE10 pKa = 4.2VLGKK14 pKa = 10.45KK15 pKa = 9.27SSKK18 pKa = 9.85SSKK21 pKa = 10.1EE22 pKa = 3.46ILGNTDD28 pKa = 3.13TKK30 pKa = 10.65ISKK33 pKa = 9.73QPSPAKK39 pKa = 10.05HH40 pKa = 5.6YY41 pKa = 11.1CFTLNNYY48 pKa = 9.78SNNEE52 pKa = 3.93LNIVPTSLGSVGHH65 pKa = 6.07YY66 pKa = 9.93IYY68 pKa = 10.79GFEE71 pKa = 4.24VGAEE75 pKa = 4.28GTPHH79 pKa = 5.53LQGYY83 pKa = 9.44IEE85 pKa = 4.73LHH87 pKa = 5.22HH88 pKa = 6.88KK89 pKa = 9.55MRR91 pKa = 11.84ITEE94 pKa = 4.15FKK96 pKa = 10.4KK97 pKa = 10.32IEE99 pKa = 4.22GLGRR103 pKa = 11.84THH105 pKa = 7.59FEE107 pKa = 3.91KK108 pKa = 10.96CKK110 pKa = 10.58GSQADD115 pKa = 3.47NLKK118 pKa = 9.77YY119 pKa = 10.09CRR121 pKa = 11.84KK122 pKa = 8.59GGNYY126 pKa = 9.09ISNYY130 pKa = 8.58WEE132 pKa = 4.25DD133 pKa = 3.38VQLIVPTYY141 pKa = 8.99YY142 pKa = 9.43WQLDD146 pKa = 3.62ILQVIASPAHH156 pKa = 5.84GRR158 pKa = 11.84HH159 pKa = 5.38IHH161 pKa = 5.75WFWEE165 pKa = 4.56SIGGVGKK172 pKa = 10.81SSFAKK177 pKa = 9.99YY178 pKa = 10.21LCSVHH183 pKa = 5.6NAIYY187 pKa = 9.96IDD189 pKa = 3.82EE190 pKa = 4.67GKK192 pKa = 10.64KK193 pKa = 10.09SDD195 pKa = 5.45LINIVYY201 pKa = 9.59NIPNITCRR209 pKa = 11.84SIIVIDD215 pKa = 3.71VPRR218 pKa = 11.84DD219 pKa = 3.34NGNTVSYY226 pKa = 10.62KK227 pKa = 10.61AIEE230 pKa = 4.02QIKK233 pKa = 10.84NGMICNTKK241 pKa = 10.19YY242 pKa = 8.43EE243 pKa = 4.14TGMRR247 pKa = 11.84LFNPPHH253 pKa = 7.55IIIFANMPPEE263 pKa = 4.0EE264 pKa = 4.86HH265 pKa = 7.11KK266 pKa = 11.12LSADD270 pKa = 2.89RR271 pKa = 11.84WKK273 pKa = 9.23ITEE276 pKa = 3.83ISSPP280 pKa = 3.5
MM1 pKa = 8.25DD2 pKa = 6.29DD3 pKa = 3.89YY4 pKa = 11.69LLKK7 pKa = 10.68FCEE10 pKa = 4.2VLGKK14 pKa = 10.45KK15 pKa = 9.27SSKK18 pKa = 9.85SSKK21 pKa = 10.1EE22 pKa = 3.46ILGNTDD28 pKa = 3.13TKK30 pKa = 10.65ISKK33 pKa = 9.73QPSPAKK39 pKa = 10.05HH40 pKa = 5.6YY41 pKa = 11.1CFTLNNYY48 pKa = 9.78SNNEE52 pKa = 3.93LNIVPTSLGSVGHH65 pKa = 6.07YY66 pKa = 9.93IYY68 pKa = 10.79GFEE71 pKa = 4.24VGAEE75 pKa = 4.28GTPHH79 pKa = 5.53LQGYY83 pKa = 9.44IEE85 pKa = 4.73LHH87 pKa = 5.22HH88 pKa = 6.88KK89 pKa = 9.55MRR91 pKa = 11.84ITEE94 pKa = 4.15FKK96 pKa = 10.4KK97 pKa = 10.32IEE99 pKa = 4.22GLGRR103 pKa = 11.84THH105 pKa = 7.59FEE107 pKa = 3.91KK108 pKa = 10.96CKK110 pKa = 10.58GSQADD115 pKa = 3.47NLKK118 pKa = 9.77YY119 pKa = 10.09CRR121 pKa = 11.84KK122 pKa = 8.59GGNYY126 pKa = 9.09ISNYY130 pKa = 8.58WEE132 pKa = 4.25DD133 pKa = 3.38VQLIVPTYY141 pKa = 8.99YY142 pKa = 9.43WQLDD146 pKa = 3.62ILQVIASPAHH156 pKa = 5.84GRR158 pKa = 11.84HH159 pKa = 5.38IHH161 pKa = 5.75WFWEE165 pKa = 4.56SIGGVGKK172 pKa = 10.81SSFAKK177 pKa = 9.99YY178 pKa = 10.21LCSVHH183 pKa = 5.6NAIYY187 pKa = 9.96IDD189 pKa = 3.82EE190 pKa = 4.67GKK192 pKa = 10.64KK193 pKa = 10.09SDD195 pKa = 5.45LINIVYY201 pKa = 9.59NIPNITCRR209 pKa = 11.84SIIVIDD215 pKa = 3.71VPRR218 pKa = 11.84DD219 pKa = 3.34NGNTVSYY226 pKa = 10.62KK227 pKa = 10.61AIEE230 pKa = 4.02QIKK233 pKa = 10.84NGMICNTKK241 pKa = 10.19YY242 pKa = 8.43EE243 pKa = 4.14TGMRR247 pKa = 11.84LFNPPHH253 pKa = 7.55IIIFANMPPEE263 pKa = 4.0EE264 pKa = 4.86HH265 pKa = 7.11KK266 pKa = 11.12LSADD270 pKa = 2.89RR271 pKa = 11.84WKK273 pKa = 9.23ITEE276 pKa = 3.83ISSPP280 pKa = 3.5
Molecular weight: 31.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9N6|A0A126G9N6_9VIRU Putative capsid protein OS=Lake Sarah-associated circular virus-19 OX=1685745 PE=4 SV=1
MM1 pKa = 8.02PLRR4 pKa = 11.84KK5 pKa = 9.37KK6 pKa = 10.14VFKK9 pKa = 10.2KK10 pKa = 10.43KK11 pKa = 10.31RR12 pKa = 11.84MPKK15 pKa = 9.68RR16 pKa = 11.84KK17 pKa = 8.75PRR19 pKa = 11.84IPKK22 pKa = 9.79GIKK25 pKa = 9.65SYY27 pKa = 10.38ISKK30 pKa = 9.15TIRR33 pKa = 11.84SNEE36 pKa = 3.94EE37 pKa = 3.57TKK39 pKa = 10.07LTSVTAGFTGYY50 pKa = 10.25NSSISSVGDD59 pKa = 3.36YY60 pKa = 7.3VTCLPPVPQGTGQSQRR76 pKa = 11.84IGQAIRR82 pKa = 11.84PIKK85 pKa = 10.22LVIRR89 pKa = 11.84GYY91 pKa = 10.72VIYY94 pKa = 10.87SADD97 pKa = 3.48SQLNARR103 pKa = 11.84MIGGRR108 pKa = 11.84MFCFSDD114 pKa = 3.43KK115 pKa = 10.43TVSNYY120 pKa = 9.62GVATSAGANYY130 pKa = 10.56NLLDD134 pKa = 3.9VGGTSQTFDD143 pKa = 3.31GTVARR148 pKa = 11.84YY149 pKa = 7.49EE150 pKa = 4.15YY151 pKa = 10.18PHH153 pKa = 7.12NNDD156 pKa = 2.52QFKK159 pKa = 10.58FYY161 pKa = 10.12IDD163 pKa = 3.03KK164 pKa = 10.19KK165 pKa = 10.0FRR167 pKa = 11.84MMKK170 pKa = 8.9PWGYY174 pKa = 8.65TNVGSTSTQAIVSMDD189 pKa = 2.72KK190 pKa = 11.07SMYY193 pKa = 10.17RR194 pKa = 11.84PFTITLTQKK203 pKa = 10.18QLPAILKK210 pKa = 9.76YY211 pKa = 10.44DD212 pKa = 3.27QQLNVNFPTNFAPYY226 pKa = 9.66VALGYY231 pKa = 10.29CDD233 pKa = 4.61LLSYY237 pKa = 10.19PADD240 pKa = 3.49TVVTQLNMTFTSTLYY255 pKa = 10.86FKK257 pKa = 10.85DD258 pKa = 3.3AA259 pKa = 4.05
MM1 pKa = 8.02PLRR4 pKa = 11.84KK5 pKa = 9.37KK6 pKa = 10.14VFKK9 pKa = 10.2KK10 pKa = 10.43KK11 pKa = 10.31RR12 pKa = 11.84MPKK15 pKa = 9.68RR16 pKa = 11.84KK17 pKa = 8.75PRR19 pKa = 11.84IPKK22 pKa = 9.79GIKK25 pKa = 9.65SYY27 pKa = 10.38ISKK30 pKa = 9.15TIRR33 pKa = 11.84SNEE36 pKa = 3.94EE37 pKa = 3.57TKK39 pKa = 10.07LTSVTAGFTGYY50 pKa = 10.25NSSISSVGDD59 pKa = 3.36YY60 pKa = 7.3VTCLPPVPQGTGQSQRR76 pKa = 11.84IGQAIRR82 pKa = 11.84PIKK85 pKa = 10.22LVIRR89 pKa = 11.84GYY91 pKa = 10.72VIYY94 pKa = 10.87SADD97 pKa = 3.48SQLNARR103 pKa = 11.84MIGGRR108 pKa = 11.84MFCFSDD114 pKa = 3.43KK115 pKa = 10.43TVSNYY120 pKa = 9.62GVATSAGANYY130 pKa = 10.56NLLDD134 pKa = 3.9VGGTSQTFDD143 pKa = 3.31GTVARR148 pKa = 11.84YY149 pKa = 7.49EE150 pKa = 4.15YY151 pKa = 10.18PHH153 pKa = 7.12NNDD156 pKa = 2.52QFKK159 pKa = 10.58FYY161 pKa = 10.12IDD163 pKa = 3.03KK164 pKa = 10.19KK165 pKa = 10.0FRR167 pKa = 11.84MMKK170 pKa = 8.9PWGYY174 pKa = 8.65TNVGSTSTQAIVSMDD189 pKa = 2.72KK190 pKa = 11.07SMYY193 pKa = 10.17RR194 pKa = 11.84PFTITLTQKK203 pKa = 10.18QLPAILKK210 pKa = 9.76YY211 pKa = 10.44DD212 pKa = 3.27QQLNVNFPTNFAPYY226 pKa = 9.66VALGYY231 pKa = 10.29CDD233 pKa = 4.61LLSYY237 pKa = 10.19PADD240 pKa = 3.49TVVTQLNMTFTSTLYY255 pKa = 10.86FKK257 pKa = 10.85DD258 pKa = 3.3AA259 pKa = 4.05
Molecular weight: 29.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
539 |
259 |
280 |
269.5 |
30.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.453 ± 0.654 | 1.855 ± 0.478 |
4.267 ± 0.251 | 3.711 ± 1.751 |
4.082 ± 0.643 | 7.236 ± 0.196 |
2.412 ± 1.39 | 8.349 ± 1.755 |
8.349 ± 0.165 | 6.308 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.597 ± 0.602 | 5.937 ± 0.63 |
5.195 ± 0.409 | 3.711 ± 0.898 |
3.896 ± 0.771 | 7.978 ± 0.089 |
6.865 ± 1.648 | 5.566 ± 0.685 |
1.113 ± 0.499 | 6.122 ± 0.303 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
