
Ancylomarina euxinus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Marinifilaceae; Ancylomarina
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
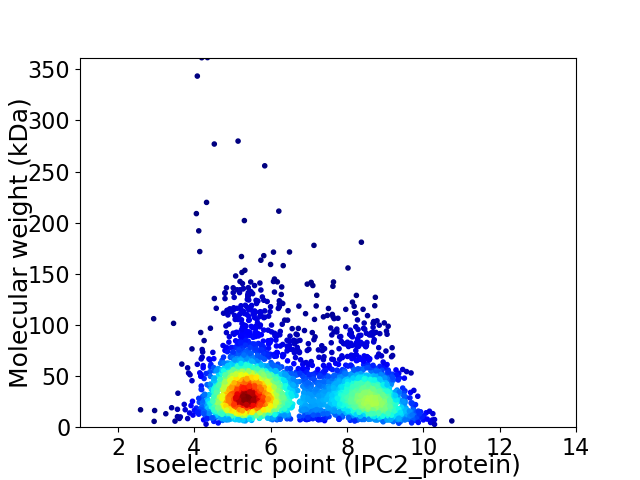
Virtual 2D-PAGE plot for 3457 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A425Y4A8|A0A425Y4A8_9BACT ATP phosphoribosyltransferase OS=Ancylomarina euxinus OX=2283627 GN=hisG PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 10.33KK3 pKa = 9.48ILYY6 pKa = 9.54FILLLVVAVSCSPDD20 pKa = 3.32EE21 pKa = 4.22YY22 pKa = 10.68STPEE26 pKa = 4.01NLTAEE31 pKa = 4.33QIDD34 pKa = 3.65WGYY37 pKa = 7.84TATDD41 pKa = 3.09ITNEE45 pKa = 3.67YY46 pKa = 8.73TLYY49 pKa = 10.98NNTPGVSSTWDD60 pKa = 3.23FGNGVTQKK68 pKa = 10.41GSSVTAQYY76 pKa = 10.31TFAGTYY82 pKa = 6.88TVKK85 pKa = 10.02LTVISQGGVIILEE98 pKa = 4.27DD99 pKa = 4.47EE100 pKa = 4.22IVTTVDD106 pKa = 3.05NPSFLSGYY114 pKa = 9.86PYY116 pKa = 10.66DD117 pKa = 3.84QLVGAGEE124 pKa = 4.26QVWAVDD130 pKa = 4.86GYY132 pKa = 11.32SKK134 pKa = 11.13GAFGLGPTLANPTEE148 pKa = 3.88WHH150 pKa = 6.79ADD152 pKa = 3.2AKK154 pKa = 10.59GARR157 pKa = 11.84LDD159 pKa = 3.4KK160 pKa = 11.05GLYY163 pKa = 9.86DD164 pKa = 4.67DD165 pKa = 5.65RR166 pKa = 11.84FTFKK170 pKa = 9.53ITEE173 pKa = 4.21SGLTLTQLTNGDD185 pKa = 3.65VYY187 pKa = 11.76ANGGWAADD195 pKa = 3.85LGSTDD200 pKa = 3.3GHH202 pKa = 5.81QEE204 pKa = 3.55PDD206 pKa = 3.17GGDD209 pKa = 3.56FIMPHH214 pKa = 5.68TGGDD218 pKa = 4.14FVCTIAGEE226 pKa = 4.29KK227 pKa = 9.49LTVTGGGFLGYY238 pKa = 10.14YY239 pKa = 10.18AGAHH243 pKa = 5.77EE244 pKa = 4.49YY245 pKa = 10.59EE246 pKa = 5.21IITLTEE252 pKa = 4.38DD253 pKa = 3.41LLEE256 pKa = 4.19VAFWDD261 pKa = 4.19TNANFYY267 pKa = 10.22WFTRR271 pKa = 11.84FAPVDD276 pKa = 3.46KK277 pKa = 9.73LTPEE281 pKa = 4.18PEE283 pKa = 4.11PVVKK287 pKa = 10.25EE288 pKa = 4.21LEE290 pKa = 4.45SNDD293 pKa = 3.32IADD296 pKa = 4.08DD297 pKa = 3.99FEE299 pKa = 6.54GNGNIIWDD307 pKa = 3.71TAAIDD312 pKa = 3.93GFDD315 pKa = 4.45IIDD318 pKa = 3.85NFAPSDD324 pKa = 3.89VNSSDD329 pKa = 5.57NIIKK333 pKa = 8.28YY334 pKa = 10.15QKK336 pKa = 10.76GAGEE340 pKa = 4.19WSSVLTVLDD349 pKa = 4.15YY350 pKa = 10.96IIDD353 pKa = 4.24LSTRR357 pKa = 11.84NQFTMKK363 pKa = 10.29VFVPAFNDD371 pKa = 3.33YY372 pKa = 7.83TTVCDD377 pKa = 5.58PGTPWIADD385 pKa = 3.64HH386 pKa = 6.42NLKK389 pKa = 9.9PQIDD393 pKa = 4.01VKK395 pKa = 11.17LQDD398 pKa = 3.48SSLGGNAWQTQQVRR412 pKa = 11.84SHH414 pKa = 6.49TLTEE418 pKa = 4.12AQFGTWVEE426 pKa = 4.1LTFDD430 pKa = 3.69YY431 pKa = 11.26SDD433 pKa = 3.23VSDD436 pKa = 5.05RR437 pKa = 11.84VDD439 pKa = 3.89FDD441 pKa = 4.08QIVIQLGAEE450 pKa = 4.28GHH452 pKa = 6.42CNSGLFYY459 pKa = 10.34IDD461 pKa = 5.64DD462 pKa = 4.1FQLLPP467 pKa = 4.16
MM1 pKa = 7.52KK2 pKa = 10.33KK3 pKa = 9.48ILYY6 pKa = 9.54FILLLVVAVSCSPDD20 pKa = 3.32EE21 pKa = 4.22YY22 pKa = 10.68STPEE26 pKa = 4.01NLTAEE31 pKa = 4.33QIDD34 pKa = 3.65WGYY37 pKa = 7.84TATDD41 pKa = 3.09ITNEE45 pKa = 3.67YY46 pKa = 8.73TLYY49 pKa = 10.98NNTPGVSSTWDD60 pKa = 3.23FGNGVTQKK68 pKa = 10.41GSSVTAQYY76 pKa = 10.31TFAGTYY82 pKa = 6.88TVKK85 pKa = 10.02LTVISQGGVIILEE98 pKa = 4.27DD99 pKa = 4.47EE100 pKa = 4.22IVTTVDD106 pKa = 3.05NPSFLSGYY114 pKa = 9.86PYY116 pKa = 10.66DD117 pKa = 3.84QLVGAGEE124 pKa = 4.26QVWAVDD130 pKa = 4.86GYY132 pKa = 11.32SKK134 pKa = 11.13GAFGLGPTLANPTEE148 pKa = 3.88WHH150 pKa = 6.79ADD152 pKa = 3.2AKK154 pKa = 10.59GARR157 pKa = 11.84LDD159 pKa = 3.4KK160 pKa = 11.05GLYY163 pKa = 9.86DD164 pKa = 4.67DD165 pKa = 5.65RR166 pKa = 11.84FTFKK170 pKa = 9.53ITEE173 pKa = 4.21SGLTLTQLTNGDD185 pKa = 3.65VYY187 pKa = 11.76ANGGWAADD195 pKa = 3.85LGSTDD200 pKa = 3.3GHH202 pKa = 5.81QEE204 pKa = 3.55PDD206 pKa = 3.17GGDD209 pKa = 3.56FIMPHH214 pKa = 5.68TGGDD218 pKa = 4.14FVCTIAGEE226 pKa = 4.29KK227 pKa = 9.49LTVTGGGFLGYY238 pKa = 10.14YY239 pKa = 10.18AGAHH243 pKa = 5.77EE244 pKa = 4.49YY245 pKa = 10.59EE246 pKa = 5.21IITLTEE252 pKa = 4.38DD253 pKa = 3.41LLEE256 pKa = 4.19VAFWDD261 pKa = 4.19TNANFYY267 pKa = 10.22WFTRR271 pKa = 11.84FAPVDD276 pKa = 3.46KK277 pKa = 9.73LTPEE281 pKa = 4.18PEE283 pKa = 4.11PVVKK287 pKa = 10.25EE288 pKa = 4.21LEE290 pKa = 4.45SNDD293 pKa = 3.32IADD296 pKa = 4.08DD297 pKa = 3.99FEE299 pKa = 6.54GNGNIIWDD307 pKa = 3.71TAAIDD312 pKa = 3.93GFDD315 pKa = 4.45IIDD318 pKa = 3.85NFAPSDD324 pKa = 3.89VNSSDD329 pKa = 5.57NIIKK333 pKa = 8.28YY334 pKa = 10.15QKK336 pKa = 10.76GAGEE340 pKa = 4.19WSSVLTVLDD349 pKa = 4.15YY350 pKa = 10.96IIDD353 pKa = 4.24LSTRR357 pKa = 11.84NQFTMKK363 pKa = 10.29VFVPAFNDD371 pKa = 3.33YY372 pKa = 7.83TTVCDD377 pKa = 5.58PGTPWIADD385 pKa = 3.64HH386 pKa = 6.42NLKK389 pKa = 9.9PQIDD393 pKa = 4.01VKK395 pKa = 11.17LQDD398 pKa = 3.48SSLGGNAWQTQQVRR412 pKa = 11.84SHH414 pKa = 6.49TLTEE418 pKa = 4.12AQFGTWVEE426 pKa = 4.1LTFDD430 pKa = 3.69YY431 pKa = 11.26SDD433 pKa = 3.23VSDD436 pKa = 5.05RR437 pKa = 11.84VDD439 pKa = 3.89FDD441 pKa = 4.08QIVIQLGAEE450 pKa = 4.28GHH452 pKa = 6.42CNSGLFYY459 pKa = 10.34IDD461 pKa = 5.64DD462 pKa = 4.1FQLLPP467 pKa = 4.16
Molecular weight: 51.28 kDa
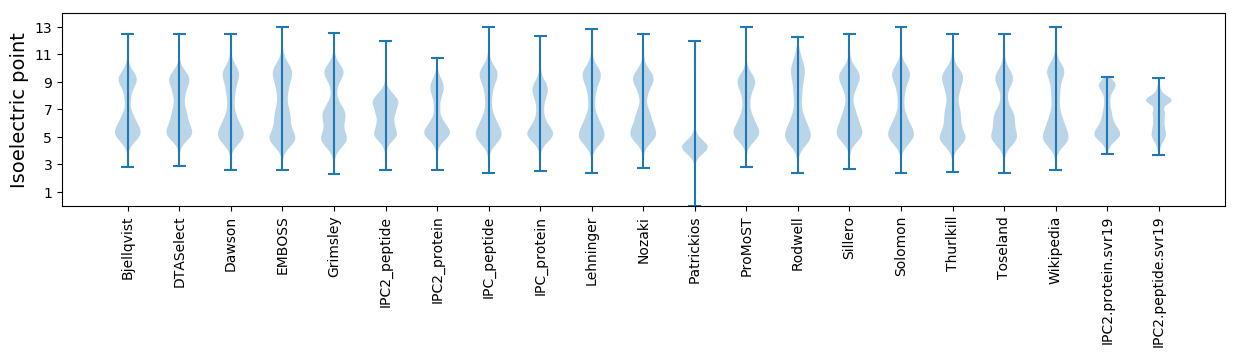
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A425Y5P2|A0A425Y5P2_9BACT Type IX secretion system membrane protein PorP/SprF OS=Ancylomarina euxinus OX=2283627 GN=DWB61_04540 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 10.23HH16 pKa = 3.88GFRR19 pKa = 11.84DD20 pKa = 3.68KK21 pKa = 10.42MASANGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.62LSVSSEE47 pKa = 3.87RR48 pKa = 11.84RR49 pKa = 11.84HH50 pKa = 5.76KK51 pKa = 10.9AA52 pKa = 2.86
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84KK11 pKa = 8.89RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 10.23HH16 pKa = 3.88GFRR19 pKa = 11.84DD20 pKa = 3.68KK21 pKa = 10.42MASANGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.14GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.62LSVSSEE47 pKa = 3.87RR48 pKa = 11.84RR49 pKa = 11.84HH50 pKa = 5.76KK51 pKa = 10.9AA52 pKa = 2.86
Molecular weight: 6.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1249026 |
23 |
3318 |
361.3 |
40.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.308 ± 0.039 | 0.988 ± 0.017 |
5.708 ± 0.031 | 6.75 ± 0.036 |
5.061 ± 0.03 | 6.311 ± 0.042 |
1.84 ± 0.016 | 8.113 ± 0.036 |
7.878 ± 0.042 | 9.622 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.482 ± 0.022 | 5.764 ± 0.035 |
3.21 ± 0.021 | 3.366 ± 0.021 |
3.699 ± 0.025 | 6.856 ± 0.036 |
5.003 ± 0.033 | 5.961 ± 0.036 |
1.049 ± 0.012 | 4.03 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
