
Apis mellifera associated microvirus 20
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.29
Get precalculated fractions of proteins
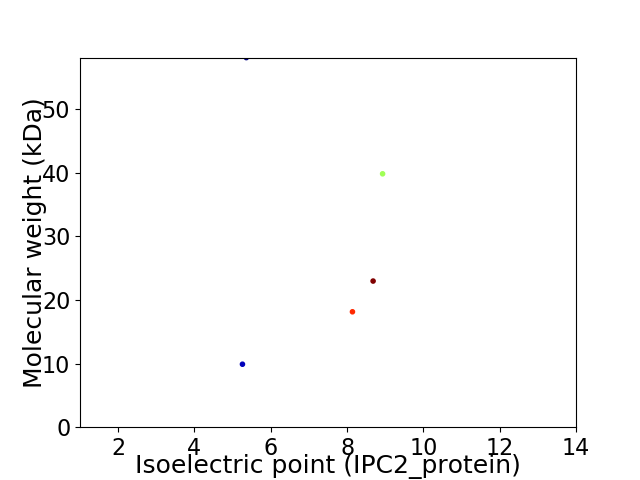
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTI4|A0A3S8UTI4_9VIRU Nonstructural protein OS=Apis mellifera associated microvirus 20 OX=2494748 PE=4 SV=1
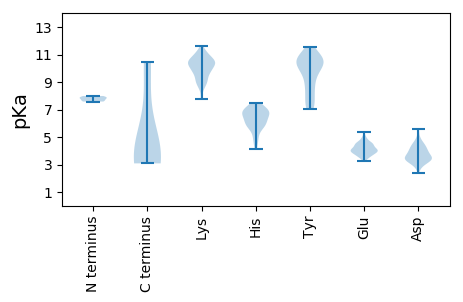
MM1 pKa = 7.63LLHH4 pKa = 7.14AFTVFDD10 pKa = 4.58SKK12 pKa = 11.28SKK14 pKa = 10.64SYY16 pKa = 8.21NTPWFARR23 pKa = 11.84SKK25 pKa = 10.95PEE27 pKa = 3.55AEE29 pKa = 4.0RR30 pKa = 11.84NFVQLVKK37 pKa = 10.74DD38 pKa = 3.98EE39 pKa = 4.54KK40 pKa = 11.61SMVNQFPEE48 pKa = 4.65DD49 pKa = 3.35FDD51 pKa = 4.24LFYY54 pKa = 10.78IGTYY58 pKa = 10.58DD59 pKa = 4.57DD60 pKa = 3.64NTAKK64 pKa = 10.3FDD66 pKa = 4.05LLEE69 pKa = 4.52TPEE72 pKa = 4.51HH73 pKa = 5.84MFKK76 pKa = 11.01ALTLKK81 pKa = 10.46NQSS84 pKa = 3.08
MM1 pKa = 7.63LLHH4 pKa = 7.14AFTVFDD10 pKa = 4.58SKK12 pKa = 11.28SKK14 pKa = 10.64SYY16 pKa = 8.21NTPWFARR23 pKa = 11.84SKK25 pKa = 10.95PEE27 pKa = 3.55AEE29 pKa = 4.0RR30 pKa = 11.84NFVQLVKK37 pKa = 10.74DD38 pKa = 3.98EE39 pKa = 4.54KK40 pKa = 11.61SMVNQFPEE48 pKa = 4.65DD49 pKa = 3.35FDD51 pKa = 4.24LFYY54 pKa = 10.78IGTYY58 pKa = 10.58DD59 pKa = 4.57DD60 pKa = 3.64NTAKK64 pKa = 10.3FDD66 pKa = 4.05LLEE69 pKa = 4.52TPEE72 pKa = 4.51HH73 pKa = 5.84MFKK76 pKa = 11.01ALTLKK81 pKa = 10.46NQSS84 pKa = 3.08
Molecular weight: 9.9 kDa
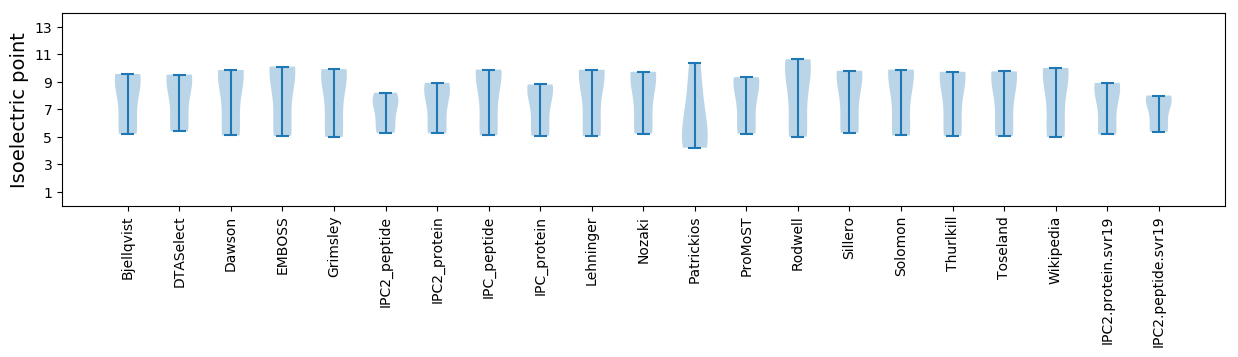
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTH3|A0A3S8UTH3_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 20 OX=2494748 PE=3 SV=1
MM1 pKa = 7.86EE2 pKa = 4.75STLLTPSTHH11 pKa = 6.34ALCVVEE17 pKa = 4.57SASNKK22 pKa = 9.83KK23 pKa = 9.77KK24 pKa = 10.29GHH26 pKa = 4.89TMRR29 pKa = 11.84CTSPRR34 pKa = 11.84TVGYY38 pKa = 9.86QADD41 pKa = 4.27GSSLSWSPKK50 pKa = 8.54TFSKK54 pKa = 10.61EE55 pKa = 3.62YY56 pKa = 9.86ATFQLPCGKK65 pKa = 9.87CLSCRR70 pKa = 11.84LEE72 pKa = 4.05YY73 pKa = 10.58ARR75 pKa = 11.84QWAVRR80 pKa = 11.84CVHH83 pKa = 6.75EE84 pKa = 5.05SMCHH88 pKa = 5.12QEE90 pKa = 3.8NCFITLTYY98 pKa = 10.58SDD100 pKa = 5.2EE101 pKa = 4.11NLKK104 pKa = 10.41SDD106 pKa = 3.29KK107 pKa = 10.69LQYY110 pKa = 10.51RR111 pKa = 11.84DD112 pKa = 3.54FQLFMKK118 pKa = 10.18RR119 pKa = 11.84LRR121 pKa = 11.84KK122 pKa = 9.65AFPNKK127 pKa = 10.07EE128 pKa = 3.61IGFNAVGEE136 pKa = 4.43YY137 pKa = 10.7GDD139 pKa = 3.71KK140 pKa = 10.34TKK142 pKa = 10.65RR143 pKa = 11.84PHH145 pKa = 4.14WHH147 pKa = 6.91AIIFNWQPSDD157 pKa = 3.44SQYY160 pKa = 10.71RR161 pKa = 11.84RR162 pKa = 11.84SNEE165 pKa = 3.44RR166 pKa = 11.84GDD168 pKa = 4.52RR169 pKa = 11.84IFSSKK174 pKa = 9.53TLDD177 pKa = 3.64KK178 pKa = 11.07LWGLGITEE186 pKa = 4.58FGSVTFHH193 pKa = 6.15SAGYY197 pKa = 8.55VSRR200 pKa = 11.84YY201 pKa = 7.14NAKK204 pKa = 10.33KK205 pKa = 10.24LIHH208 pKa = 6.69GKK210 pKa = 8.84DD211 pKa = 3.55QEE213 pKa = 4.64HH214 pKa = 7.3DD215 pKa = 3.42LHH217 pKa = 6.78PISKK221 pKa = 9.98KK222 pKa = 9.09SNKK225 pKa = 9.02HH226 pKa = 5.82AIGKK230 pKa = 9.0KK231 pKa = 7.81WLEE234 pKa = 4.58KK235 pKa = 9.76YY236 pKa = 8.98WQDD239 pKa = 3.28AFNHH243 pKa = 6.06GEE245 pKa = 4.14IILADD250 pKa = 3.94GTSTAIPRR258 pKa = 11.84YY259 pKa = 7.05YY260 pKa = 10.54EE261 pKa = 3.61KK262 pKa = 10.07WLKK265 pKa = 10.14EE266 pKa = 4.18NQPEE270 pKa = 4.02AYY272 pKa = 9.59LRR274 pKa = 11.84YY275 pKa = 8.68VTQTKK280 pKa = 9.32IRR282 pKa = 11.84KK283 pKa = 8.91IEE285 pKa = 3.99AAQEE289 pKa = 4.03KK290 pKa = 9.29AAKK293 pKa = 8.97EE294 pKa = 3.85ATRR297 pKa = 11.84AEE299 pKa = 4.26LANHH303 pKa = 6.29KK304 pKa = 10.3RR305 pKa = 11.84RR306 pKa = 11.84TASLLDD312 pKa = 4.41GYY314 pKa = 11.1HH315 pKa = 6.82RR316 pKa = 11.84GHH318 pKa = 6.7EE319 pKa = 3.91ISRR322 pKa = 11.84NEE324 pKa = 3.78INKK327 pKa = 9.87KK328 pKa = 8.45IQQARR333 pKa = 11.84FKK335 pKa = 10.73QLQDD339 pKa = 3.34NLKK342 pKa = 10.42LL343 pKa = 3.74
MM1 pKa = 7.86EE2 pKa = 4.75STLLTPSTHH11 pKa = 6.34ALCVVEE17 pKa = 4.57SASNKK22 pKa = 9.83KK23 pKa = 9.77KK24 pKa = 10.29GHH26 pKa = 4.89TMRR29 pKa = 11.84CTSPRR34 pKa = 11.84TVGYY38 pKa = 9.86QADD41 pKa = 4.27GSSLSWSPKK50 pKa = 8.54TFSKK54 pKa = 10.61EE55 pKa = 3.62YY56 pKa = 9.86ATFQLPCGKK65 pKa = 9.87CLSCRR70 pKa = 11.84LEE72 pKa = 4.05YY73 pKa = 10.58ARR75 pKa = 11.84QWAVRR80 pKa = 11.84CVHH83 pKa = 6.75EE84 pKa = 5.05SMCHH88 pKa = 5.12QEE90 pKa = 3.8NCFITLTYY98 pKa = 10.58SDD100 pKa = 5.2EE101 pKa = 4.11NLKK104 pKa = 10.41SDD106 pKa = 3.29KK107 pKa = 10.69LQYY110 pKa = 10.51RR111 pKa = 11.84DD112 pKa = 3.54FQLFMKK118 pKa = 10.18RR119 pKa = 11.84LRR121 pKa = 11.84KK122 pKa = 9.65AFPNKK127 pKa = 10.07EE128 pKa = 3.61IGFNAVGEE136 pKa = 4.43YY137 pKa = 10.7GDD139 pKa = 3.71KK140 pKa = 10.34TKK142 pKa = 10.65RR143 pKa = 11.84PHH145 pKa = 4.14WHH147 pKa = 6.91AIIFNWQPSDD157 pKa = 3.44SQYY160 pKa = 10.71RR161 pKa = 11.84RR162 pKa = 11.84SNEE165 pKa = 3.44RR166 pKa = 11.84GDD168 pKa = 4.52RR169 pKa = 11.84IFSSKK174 pKa = 9.53TLDD177 pKa = 3.64KK178 pKa = 11.07LWGLGITEE186 pKa = 4.58FGSVTFHH193 pKa = 6.15SAGYY197 pKa = 8.55VSRR200 pKa = 11.84YY201 pKa = 7.14NAKK204 pKa = 10.33KK205 pKa = 10.24LIHH208 pKa = 6.69GKK210 pKa = 8.84DD211 pKa = 3.55QEE213 pKa = 4.64HH214 pKa = 7.3DD215 pKa = 3.42LHH217 pKa = 6.78PISKK221 pKa = 9.98KK222 pKa = 9.09SNKK225 pKa = 9.02HH226 pKa = 5.82AIGKK230 pKa = 9.0KK231 pKa = 7.81WLEE234 pKa = 4.58KK235 pKa = 9.76YY236 pKa = 8.98WQDD239 pKa = 3.28AFNHH243 pKa = 6.06GEE245 pKa = 4.14IILADD250 pKa = 3.94GTSTAIPRR258 pKa = 11.84YY259 pKa = 7.05YY260 pKa = 10.54EE261 pKa = 3.61KK262 pKa = 10.07WLKK265 pKa = 10.14EE266 pKa = 4.18NQPEE270 pKa = 4.02AYY272 pKa = 9.59LRR274 pKa = 11.84YY275 pKa = 8.68VTQTKK280 pKa = 9.32IRR282 pKa = 11.84KK283 pKa = 8.91IEE285 pKa = 3.99AAQEE289 pKa = 4.03KK290 pKa = 9.29AAKK293 pKa = 8.97EE294 pKa = 3.85ATRR297 pKa = 11.84AEE299 pKa = 4.26LANHH303 pKa = 6.29KK304 pKa = 10.3RR305 pKa = 11.84RR306 pKa = 11.84TASLLDD312 pKa = 4.41GYY314 pKa = 11.1HH315 pKa = 6.82RR316 pKa = 11.84GHH318 pKa = 6.7EE319 pKa = 3.91ISRR322 pKa = 11.84NEE324 pKa = 3.78INKK327 pKa = 9.87KK328 pKa = 8.45IQQARR333 pKa = 11.84FKK335 pKa = 10.73QLQDD339 pKa = 3.34NLKK342 pKa = 10.42LL343 pKa = 3.74
Molecular weight: 39.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1320 |
84 |
520 |
264.0 |
29.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.561 ± 1.729 | 0.758 ± 0.49 |
5.0 ± 0.809 | 6.061 ± 0.842 |
4.924 ± 1.205 | 5.985 ± 0.965 |
2.576 ± 0.653 | 5.455 ± 0.629 |
6.97 ± 1.963 | 7.424 ± 0.804 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.333 ± 1.045 | 5.833 ± 0.79 |
4.848 ± 0.616 | 5.0 ± 0.818 |
5.455 ± 0.856 | 6.439 ± 0.524 |
6.667 ± 0.888 | 4.47 ± 0.579 |
1.288 ± 0.474 | 2.955 ± 0.667 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
