
Fowl aviadenovirus E
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Aviadenovirus
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
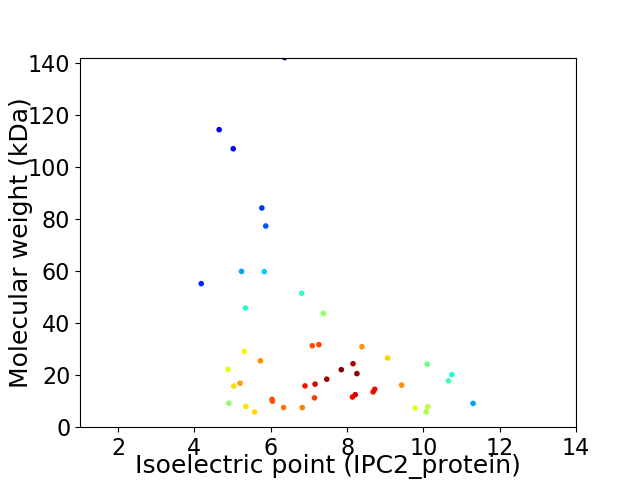
Virtual 2D-PAGE plot for 46 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E9KLB4|E9KLB4_9ADEN ORF32 OS=Fowl aviadenovirus E OX=190065 PE=4 SV=1
MM1 pKa = 7.12ATSTPHH7 pKa = 6.67AFSFGQIGSRR17 pKa = 11.84KK18 pKa = 9.34RR19 pKa = 11.84PAGGDD24 pKa = 3.67GEE26 pKa = 4.79RR27 pKa = 11.84DD28 pKa = 3.22ASKK31 pKa = 10.42VPKK34 pKa = 8.97MQTPAPSATANGNDD48 pKa = 4.13EE49 pKa = 5.04LDD51 pKa = 3.4LVYY54 pKa = 10.27PFWLQNGSTGGGGGGGGSGGNPSLNPPFLDD84 pKa = 3.61PNGPLAVQNNLLKK97 pKa = 11.07VNTAAPITVANKK109 pKa = 10.35ALTLAYY115 pKa = 9.96EE116 pKa = 4.36PDD118 pKa = 3.78SLEE121 pKa = 4.31LTNQQQLAVKK131 pKa = 9.52IDD133 pKa = 3.54PEE135 pKa = 5.08GPLKK139 pKa = 10.43ATTEE143 pKa = 4.8GIQLSVDD150 pKa = 3.45PTTLEE155 pKa = 3.71VDD157 pKa = 3.65DD158 pKa = 5.05VDD160 pKa = 3.82WEE162 pKa = 4.5LTVKK166 pKa = 10.64LDD168 pKa = 3.77PDD170 pKa = 4.22GPLDD174 pKa = 3.87SSATGITVRR183 pKa = 11.84VDD185 pKa = 3.12EE186 pKa = 4.58TLLIEE191 pKa = 5.21DD192 pKa = 4.14VGSGQGKK199 pKa = 9.0EE200 pKa = 3.96LGVNLNPTGPITADD214 pKa = 3.45DD215 pKa = 3.66QGLDD219 pKa = 4.5LEE221 pKa = 5.33IDD223 pKa = 3.64NQTLKK228 pKa = 10.94VNSVTGGGVLAVQLKK243 pKa = 9.59SQGGLTAQTDD253 pKa = 5.39GIQVNTQNSITVTNGALDD271 pKa = 3.61VKK273 pKa = 10.68VAANGPLEE281 pKa = 4.23STDD284 pKa = 3.14TGLTLNYY291 pKa = 10.33DD292 pKa = 3.79PGDD295 pKa = 3.62FTVNAGTLSIIRR307 pKa = 11.84DD308 pKa = 3.59PALVANAYY316 pKa = 7.16LTSGASTLQQFTAKK330 pKa = 10.14SEE332 pKa = 4.06NSSQFSFPCAYY343 pKa = 10.45YY344 pKa = 9.99LQQWLSDD351 pKa = 3.64GLVLSSLYY359 pKa = 10.79LKK361 pKa = 10.5LDD363 pKa = 3.56RR364 pKa = 11.84AQFTNMPTGANYY376 pKa = 10.13QNARR380 pKa = 11.84YY381 pKa = 7.53FTFWVGAGTSFNLSTLTEE399 pKa = 4.02PTITPNTTQWNAFAPALDD417 pKa = 4.12YY418 pKa = 11.51SGAPPFIYY426 pKa = 10.17DD427 pKa = 3.38ASSVVTIYY435 pKa = 10.55FEE437 pKa = 4.19PTSGRR442 pKa = 11.84LEE444 pKa = 4.22SYY446 pKa = 10.98LPVLTDD452 pKa = 3.12NWSQTYY458 pKa = 10.61NPGTVTLCVKK468 pKa = 8.17TVRR471 pKa = 11.84VQLRR475 pKa = 11.84SQGTFSTLVCYY486 pKa = 10.21NFRR489 pKa = 11.84CQNTGIFNSNATAGTMTLGPIFFSCPALSTANAPP523 pKa = 3.63
MM1 pKa = 7.12ATSTPHH7 pKa = 6.67AFSFGQIGSRR17 pKa = 11.84KK18 pKa = 9.34RR19 pKa = 11.84PAGGDD24 pKa = 3.67GEE26 pKa = 4.79RR27 pKa = 11.84DD28 pKa = 3.22ASKK31 pKa = 10.42VPKK34 pKa = 8.97MQTPAPSATANGNDD48 pKa = 4.13EE49 pKa = 5.04LDD51 pKa = 3.4LVYY54 pKa = 10.27PFWLQNGSTGGGGGGGGSGGNPSLNPPFLDD84 pKa = 3.61PNGPLAVQNNLLKK97 pKa = 11.07VNTAAPITVANKK109 pKa = 10.35ALTLAYY115 pKa = 9.96EE116 pKa = 4.36PDD118 pKa = 3.78SLEE121 pKa = 4.31LTNQQQLAVKK131 pKa = 9.52IDD133 pKa = 3.54PEE135 pKa = 5.08GPLKK139 pKa = 10.43ATTEE143 pKa = 4.8GIQLSVDD150 pKa = 3.45PTTLEE155 pKa = 3.71VDD157 pKa = 3.65DD158 pKa = 5.05VDD160 pKa = 3.82WEE162 pKa = 4.5LTVKK166 pKa = 10.64LDD168 pKa = 3.77PDD170 pKa = 4.22GPLDD174 pKa = 3.87SSATGITVRR183 pKa = 11.84VDD185 pKa = 3.12EE186 pKa = 4.58TLLIEE191 pKa = 5.21DD192 pKa = 4.14VGSGQGKK199 pKa = 9.0EE200 pKa = 3.96LGVNLNPTGPITADD214 pKa = 3.45DD215 pKa = 3.66QGLDD219 pKa = 4.5LEE221 pKa = 5.33IDD223 pKa = 3.64NQTLKK228 pKa = 10.94VNSVTGGGVLAVQLKK243 pKa = 9.59SQGGLTAQTDD253 pKa = 5.39GIQVNTQNSITVTNGALDD271 pKa = 3.61VKK273 pKa = 10.68VAANGPLEE281 pKa = 4.23STDD284 pKa = 3.14TGLTLNYY291 pKa = 10.33DD292 pKa = 3.79PGDD295 pKa = 3.62FTVNAGTLSIIRR307 pKa = 11.84DD308 pKa = 3.59PALVANAYY316 pKa = 7.16LTSGASTLQQFTAKK330 pKa = 10.14SEE332 pKa = 4.06NSSQFSFPCAYY343 pKa = 10.45YY344 pKa = 9.99LQQWLSDD351 pKa = 3.64GLVLSSLYY359 pKa = 10.79LKK361 pKa = 10.5LDD363 pKa = 3.56RR364 pKa = 11.84AQFTNMPTGANYY376 pKa = 10.13QNARR380 pKa = 11.84YY381 pKa = 7.53FTFWVGAGTSFNLSTLTEE399 pKa = 4.02PTITPNTTQWNAFAPALDD417 pKa = 4.12YY418 pKa = 11.51SGAPPFIYY426 pKa = 10.17DD427 pKa = 3.38ASSVVTIYY435 pKa = 10.55FEE437 pKa = 4.19PTSGRR442 pKa = 11.84LEE444 pKa = 4.22SYY446 pKa = 10.98LPVLTDD452 pKa = 3.12NWSQTYY458 pKa = 10.61NPGTVTLCVKK468 pKa = 8.17TVRR471 pKa = 11.84VQLRR475 pKa = 11.84SQGTFSTLVCYY486 pKa = 10.21NFRR489 pKa = 11.84CQNTGIFNSNATAGTMTLGPIFFSCPALSTANAPP523 pKa = 3.63
Molecular weight: 55.13 kDa
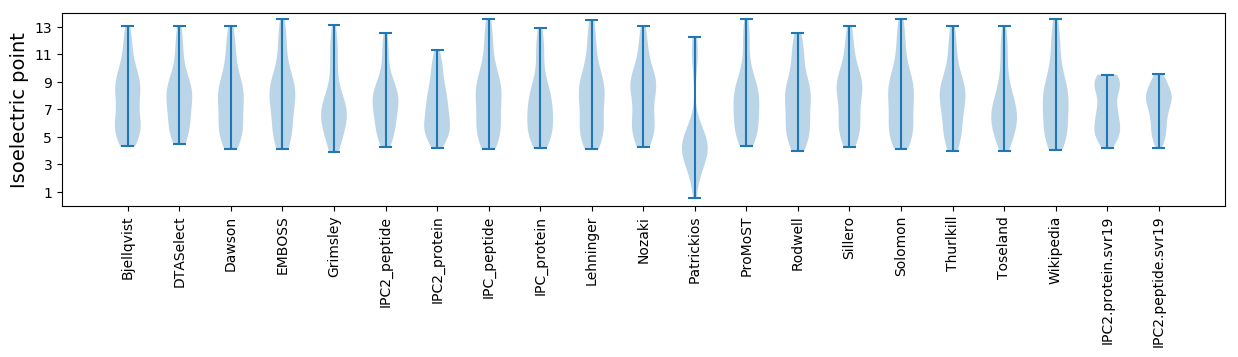
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E9KLA4|E9KLA4_9ADEN Core protein pX OS=Fowl aviadenovirus E OX=190065 PE=4 SV=1
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LTLRR36 pKa = 11.84TLLGLGTVSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GGRR52 pKa = 11.84SSRR55 pKa = 11.84RR56 pKa = 11.84SSRR59 pKa = 11.84PASTTTRR66 pKa = 11.84LMVVRR71 pKa = 11.84TSRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 3.32
MM1 pKa = 7.68SILISPNDD9 pKa = 3.25NRR11 pKa = 11.84GWGMRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84SSMRR24 pKa = 11.84GVGIRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84LTLRR36 pKa = 11.84TLLGLGTVSRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84GGRR52 pKa = 11.84SSRR55 pKa = 11.84RR56 pKa = 11.84SSRR59 pKa = 11.84PASTTTRR66 pKa = 11.84LMVVRR71 pKa = 11.84TSRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 3.32
Molecular weight: 9.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12659 |
54 |
1226 |
275.2 |
30.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.018 ± 0.454 | 1.88 ± 0.233 |
5.419 ± 0.378 | 5.79 ± 0.436 |
4.139 ± 0.216 | 6.841 ± 0.398 |
2.401 ± 0.211 | 3.507 ± 0.15 |
3.247 ± 0.335 | 8.966 ± 0.341 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.409 ± 0.17 | 3.855 ± 0.4 |
6.896 ± 0.393 | 3.705 ± 0.233 |
8.042 ± 0.668 | 7.102 ± 0.55 |
6.019 ± 0.388 | 6.636 ± 0.345 |
1.382 ± 0.145 | 3.744 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
