
Rosellinia necatrix partitivirus 6
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus; unclassified Betapartitivirus
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
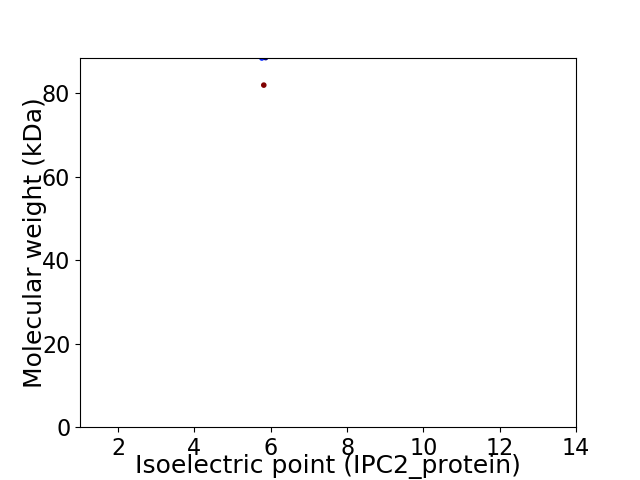
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
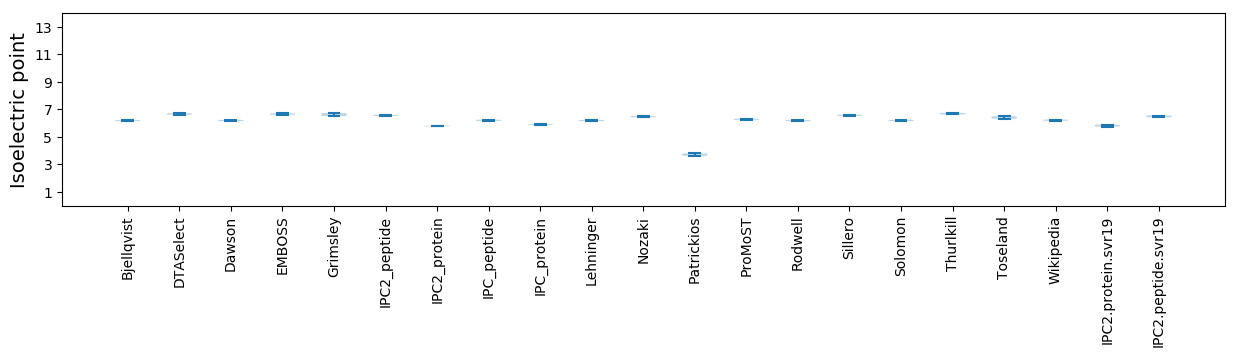
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0YTQ4|A0A0P0YTQ4_9VIRU RNA dependent RNA polymerase OS=Rosellinia necatrix partitivirus 6 OX=1573459 GN=RdRp PE=4 SV=1
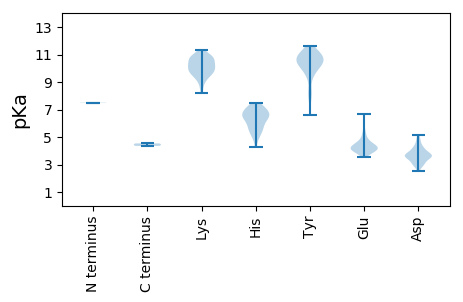
MM1 pKa = 7.47SSVFNNVRR9 pKa = 11.84NYY11 pKa = 10.01LQEE14 pKa = 4.1RR15 pKa = 11.84LQHH18 pKa = 6.52VKK20 pKa = 10.59RR21 pKa = 11.84EE22 pKa = 4.04WQIYY26 pKa = 7.4QATGASSSNMTEE38 pKa = 3.63EE39 pKa = 5.98RR40 pKa = 11.84IQQLQDD46 pKa = 2.93LDD48 pKa = 3.87TRR50 pKa = 11.84RR51 pKa = 11.84MFHH54 pKa = 5.45TARR57 pKa = 11.84NAFDD61 pKa = 3.63TTTNTPGPVSRR72 pKa = 11.84ITEE75 pKa = 3.88EE76 pKa = 3.89QRR78 pKa = 11.84RR79 pKa = 11.84LIYY82 pKa = 10.37DD83 pKa = 3.76AEE85 pKa = 4.31SDD87 pKa = 3.8KK88 pKa = 11.36IMNALRR94 pKa = 11.84QHH96 pKa = 7.1DD97 pKa = 4.26EE98 pKa = 3.85MRR100 pKa = 11.84SQPFEE105 pKa = 4.42LFMTRR110 pKa = 11.84PDD112 pKa = 3.97SDD114 pKa = 5.07PIPPNRR120 pKa = 11.84IAAPGIWQSPLQFHH134 pKa = 6.81TGQIVHH140 pKa = 7.46ADD142 pKa = 3.53PSTTSVLHH150 pKa = 6.44PDD152 pKa = 2.99EE153 pKa = 6.67HH154 pKa = 6.69EE155 pKa = 5.31DD156 pKa = 3.69YY157 pKa = 11.33AEE159 pKa = 4.43SYY161 pKa = 11.14LPGDD165 pKa = 3.29TDD167 pKa = 2.9QGYY170 pKa = 10.31EE171 pKa = 3.98IDD173 pKa = 3.63PTIYY177 pKa = 10.52EE178 pKa = 4.4LLTRR182 pKa = 11.84KK183 pKa = 9.82YY184 pKa = 10.38PEE186 pKa = 4.1YY187 pKa = 10.66LPYY190 pKa = 10.03AQQYY194 pKa = 8.22CRR196 pKa = 11.84PKK198 pKa = 9.73GTTDD202 pKa = 2.79GTFRR206 pKa = 11.84DD207 pKa = 4.32FNKK210 pKa = 9.87EE211 pKa = 3.86QKK213 pKa = 9.74PCPPLDD219 pKa = 3.57SDD221 pKa = 4.35RR222 pKa = 11.84KK223 pKa = 8.5EE224 pKa = 4.14HH225 pKa = 5.66VLKK228 pKa = 10.95HH229 pKa = 4.31VFKK232 pKa = 10.92RR233 pKa = 11.84LAIDD237 pKa = 4.35PYY239 pKa = 11.16LPLHH243 pKa = 6.44FVDD246 pKa = 3.82TQYY249 pKa = 11.64CKK251 pKa = 10.91LPLVTGTGYY260 pKa = 10.77HH261 pKa = 5.45NRR263 pKa = 11.84YY264 pKa = 9.68SYY266 pKa = 10.24RR267 pKa = 11.84QKK269 pKa = 10.09AHH271 pKa = 6.8ASFSHH276 pKa = 6.42PLQYY280 pKa = 10.89GSRR283 pKa = 11.84PTSKK287 pKa = 10.74GYY289 pKa = 10.3FYY291 pKa = 11.19NATYY295 pKa = 10.76EE296 pKa = 3.93NARR299 pKa = 11.84TIIHH303 pKa = 6.57NIKK306 pKa = 10.42EE307 pKa = 4.2SGVPFNIHH315 pKa = 6.42FAPEE319 pKa = 4.05DD320 pKa = 3.66RR321 pKa = 11.84DD322 pKa = 3.71ITDD325 pKa = 3.61TEE327 pKa = 3.57IDD329 pKa = 3.54EE330 pKa = 4.63YY331 pKa = 11.17RR332 pKa = 11.84RR333 pKa = 11.84KK334 pKa = 10.51CNNFFDD340 pKa = 4.24QYY342 pKa = 9.59PTLLFTRR349 pKa = 11.84NHH351 pKa = 5.84ISNRR355 pKa = 11.84DD356 pKa = 3.1GTLKK360 pKa = 10.22VRR362 pKa = 11.84PVYY365 pKa = 10.59AVDD368 pKa = 4.8DD369 pKa = 3.63IFIIIEE375 pKa = 4.05AMLTFPALVQARR387 pKa = 11.84KK388 pKa = 9.12PDD390 pKa = 3.61CCIMYY395 pKa = 10.41GLEE398 pKa = 4.22TIRR401 pKa = 11.84GANHH405 pKa = 6.97KK406 pKa = 9.95LDD408 pKa = 3.97SLAQSFSTYY417 pKa = 8.13FTIDD421 pKa = 2.55WSGYY425 pKa = 6.69DD426 pKa = 3.13QRR428 pKa = 11.84LPRR431 pKa = 11.84AITDD435 pKa = 3.85LFYY438 pKa = 10.74TDD440 pKa = 3.65FLRR443 pKa = 11.84RR444 pKa = 11.84LIVISHH450 pKa = 7.03GYY452 pKa = 8.33QPTYY456 pKa = 10.43EE457 pKa = 4.22YY458 pKa = 8.46PTYY461 pKa = 10.52PDD463 pKa = 3.97LNEE466 pKa = 4.66DD467 pKa = 3.38NMYY470 pKa = 11.51DD471 pKa = 5.17RR472 pKa = 11.84MDD474 pKa = 4.61KK475 pKa = 10.51LLKK478 pKa = 9.37WLHH481 pKa = 5.72LWYY484 pKa = 11.22NNMTFVTADD493 pKa = 3.54GYY495 pKa = 11.44AYY497 pKa = 10.18RR498 pKa = 11.84RR499 pKa = 11.84MYY501 pKa = 10.98AGVPSGLFNTQFLDD515 pKa = 3.59SFANLYY521 pKa = 10.27ILIDD525 pKa = 3.59GMIEE529 pKa = 3.91FGFTDD534 pKa = 3.65EE535 pKa = 5.32EE536 pKa = 4.23IDD538 pKa = 3.86SFLLFVLGDD547 pKa = 4.22DD548 pKa = 3.99NSGMTNLSLARR559 pKa = 11.84LHH561 pKa = 6.65EE562 pKa = 4.98FIQFLEE568 pKa = 4.76AYY570 pKa = 9.66ALTRR574 pKa = 11.84YY575 pKa = 10.27HH576 pKa = 6.22MVLSHH581 pKa = 6.38TKK583 pKa = 10.35SVITALRR590 pKa = 11.84NKK592 pKa = 9.77IEE594 pKa = 4.13SLGYY598 pKa = 9.25QCNFGLPKK606 pKa = 9.82RR607 pKa = 11.84DD608 pKa = 2.97IGKK611 pKa = 9.63LVAQLIYY618 pKa = 9.87PEE620 pKa = 4.9HH621 pKa = 6.8KK622 pKa = 9.87IKK624 pKa = 10.68YY625 pKa = 7.91HH626 pKa = 4.87TMSARR631 pKa = 11.84AIGLAYY637 pKa = 10.23ASCAYY642 pKa = 10.57DD643 pKa = 3.03KK644 pKa = 11.03TFYY647 pKa = 10.81NFCKK651 pKa = 10.22SIYY654 pKa = 10.37NIFLDD659 pKa = 3.92YY660 pKa = 11.37YY661 pKa = 10.63EE662 pKa = 4.84YY663 pKa = 10.89DD664 pKa = 3.42EE665 pKa = 4.52KK666 pKa = 10.76TALNLSRR673 pKa = 11.84FLTTGQDD680 pKa = 3.46DD681 pKa = 4.31LTTQFSWKK689 pKa = 9.53ILPPFPTRR697 pKa = 11.84EE698 pKa = 4.19EE699 pKa = 3.55IRR701 pKa = 11.84KK702 pKa = 9.61QISFYY707 pKa = 10.8HH708 pKa = 6.42GPLDD712 pKa = 4.94YY713 pKa = 10.97APKK716 pKa = 9.86WNFAHH721 pKa = 7.39FINKK725 pKa = 8.99PDD727 pKa = 4.0VIPPSSKK734 pKa = 9.47TMYY737 pKa = 10.16DD738 pKa = 3.47YY739 pKa = 10.95EE740 pKa = 4.68IEE742 pKa = 4.3HH743 pKa = 6.55SLRR746 pKa = 11.84PQPAPTFVARR756 pKa = 4.36
MM1 pKa = 7.47SSVFNNVRR9 pKa = 11.84NYY11 pKa = 10.01LQEE14 pKa = 4.1RR15 pKa = 11.84LQHH18 pKa = 6.52VKK20 pKa = 10.59RR21 pKa = 11.84EE22 pKa = 4.04WQIYY26 pKa = 7.4QATGASSSNMTEE38 pKa = 3.63EE39 pKa = 5.98RR40 pKa = 11.84IQQLQDD46 pKa = 2.93LDD48 pKa = 3.87TRR50 pKa = 11.84RR51 pKa = 11.84MFHH54 pKa = 5.45TARR57 pKa = 11.84NAFDD61 pKa = 3.63TTTNTPGPVSRR72 pKa = 11.84ITEE75 pKa = 3.88EE76 pKa = 3.89QRR78 pKa = 11.84RR79 pKa = 11.84LIYY82 pKa = 10.37DD83 pKa = 3.76AEE85 pKa = 4.31SDD87 pKa = 3.8KK88 pKa = 11.36IMNALRR94 pKa = 11.84QHH96 pKa = 7.1DD97 pKa = 4.26EE98 pKa = 3.85MRR100 pKa = 11.84SQPFEE105 pKa = 4.42LFMTRR110 pKa = 11.84PDD112 pKa = 3.97SDD114 pKa = 5.07PIPPNRR120 pKa = 11.84IAAPGIWQSPLQFHH134 pKa = 6.81TGQIVHH140 pKa = 7.46ADD142 pKa = 3.53PSTTSVLHH150 pKa = 6.44PDD152 pKa = 2.99EE153 pKa = 6.67HH154 pKa = 6.69EE155 pKa = 5.31DD156 pKa = 3.69YY157 pKa = 11.33AEE159 pKa = 4.43SYY161 pKa = 11.14LPGDD165 pKa = 3.29TDD167 pKa = 2.9QGYY170 pKa = 10.31EE171 pKa = 3.98IDD173 pKa = 3.63PTIYY177 pKa = 10.52EE178 pKa = 4.4LLTRR182 pKa = 11.84KK183 pKa = 9.82YY184 pKa = 10.38PEE186 pKa = 4.1YY187 pKa = 10.66LPYY190 pKa = 10.03AQQYY194 pKa = 8.22CRR196 pKa = 11.84PKK198 pKa = 9.73GTTDD202 pKa = 2.79GTFRR206 pKa = 11.84DD207 pKa = 4.32FNKK210 pKa = 9.87EE211 pKa = 3.86QKK213 pKa = 9.74PCPPLDD219 pKa = 3.57SDD221 pKa = 4.35RR222 pKa = 11.84KK223 pKa = 8.5EE224 pKa = 4.14HH225 pKa = 5.66VLKK228 pKa = 10.95HH229 pKa = 4.31VFKK232 pKa = 10.92RR233 pKa = 11.84LAIDD237 pKa = 4.35PYY239 pKa = 11.16LPLHH243 pKa = 6.44FVDD246 pKa = 3.82TQYY249 pKa = 11.64CKK251 pKa = 10.91LPLVTGTGYY260 pKa = 10.77HH261 pKa = 5.45NRR263 pKa = 11.84YY264 pKa = 9.68SYY266 pKa = 10.24RR267 pKa = 11.84QKK269 pKa = 10.09AHH271 pKa = 6.8ASFSHH276 pKa = 6.42PLQYY280 pKa = 10.89GSRR283 pKa = 11.84PTSKK287 pKa = 10.74GYY289 pKa = 10.3FYY291 pKa = 11.19NATYY295 pKa = 10.76EE296 pKa = 3.93NARR299 pKa = 11.84TIIHH303 pKa = 6.57NIKK306 pKa = 10.42EE307 pKa = 4.2SGVPFNIHH315 pKa = 6.42FAPEE319 pKa = 4.05DD320 pKa = 3.66RR321 pKa = 11.84DD322 pKa = 3.71ITDD325 pKa = 3.61TEE327 pKa = 3.57IDD329 pKa = 3.54EE330 pKa = 4.63YY331 pKa = 11.17RR332 pKa = 11.84RR333 pKa = 11.84KK334 pKa = 10.51CNNFFDD340 pKa = 4.24QYY342 pKa = 9.59PTLLFTRR349 pKa = 11.84NHH351 pKa = 5.84ISNRR355 pKa = 11.84DD356 pKa = 3.1GTLKK360 pKa = 10.22VRR362 pKa = 11.84PVYY365 pKa = 10.59AVDD368 pKa = 4.8DD369 pKa = 3.63IFIIIEE375 pKa = 4.05AMLTFPALVQARR387 pKa = 11.84KK388 pKa = 9.12PDD390 pKa = 3.61CCIMYY395 pKa = 10.41GLEE398 pKa = 4.22TIRR401 pKa = 11.84GANHH405 pKa = 6.97KK406 pKa = 9.95LDD408 pKa = 3.97SLAQSFSTYY417 pKa = 8.13FTIDD421 pKa = 2.55WSGYY425 pKa = 6.69DD426 pKa = 3.13QRR428 pKa = 11.84LPRR431 pKa = 11.84AITDD435 pKa = 3.85LFYY438 pKa = 10.74TDD440 pKa = 3.65FLRR443 pKa = 11.84RR444 pKa = 11.84LIVISHH450 pKa = 7.03GYY452 pKa = 8.33QPTYY456 pKa = 10.43EE457 pKa = 4.22YY458 pKa = 8.46PTYY461 pKa = 10.52PDD463 pKa = 3.97LNEE466 pKa = 4.66DD467 pKa = 3.38NMYY470 pKa = 11.51DD471 pKa = 5.17RR472 pKa = 11.84MDD474 pKa = 4.61KK475 pKa = 10.51LLKK478 pKa = 9.37WLHH481 pKa = 5.72LWYY484 pKa = 11.22NNMTFVTADD493 pKa = 3.54GYY495 pKa = 11.44AYY497 pKa = 10.18RR498 pKa = 11.84RR499 pKa = 11.84MYY501 pKa = 10.98AGVPSGLFNTQFLDD515 pKa = 3.59SFANLYY521 pKa = 10.27ILIDD525 pKa = 3.59GMIEE529 pKa = 3.91FGFTDD534 pKa = 3.65EE535 pKa = 5.32EE536 pKa = 4.23IDD538 pKa = 3.86SFLLFVLGDD547 pKa = 4.22DD548 pKa = 3.99NSGMTNLSLARR559 pKa = 11.84LHH561 pKa = 6.65EE562 pKa = 4.98FIQFLEE568 pKa = 4.76AYY570 pKa = 9.66ALTRR574 pKa = 11.84YY575 pKa = 10.27HH576 pKa = 6.22MVLSHH581 pKa = 6.38TKK583 pKa = 10.35SVITALRR590 pKa = 11.84NKK592 pKa = 9.77IEE594 pKa = 4.13SLGYY598 pKa = 9.25QCNFGLPKK606 pKa = 9.82RR607 pKa = 11.84DD608 pKa = 2.97IGKK611 pKa = 9.63LVAQLIYY618 pKa = 9.87PEE620 pKa = 4.9HH621 pKa = 6.8KK622 pKa = 9.87IKK624 pKa = 10.68YY625 pKa = 7.91HH626 pKa = 4.87TMSARR631 pKa = 11.84AIGLAYY637 pKa = 10.23ASCAYY642 pKa = 10.57DD643 pKa = 3.03KK644 pKa = 11.03TFYY647 pKa = 10.81NFCKK651 pKa = 10.22SIYY654 pKa = 10.37NIFLDD659 pKa = 3.92YY660 pKa = 11.37YY661 pKa = 10.63EE662 pKa = 4.84YY663 pKa = 10.89DD664 pKa = 3.42EE665 pKa = 4.52KK666 pKa = 10.76TALNLSRR673 pKa = 11.84FLTTGQDD680 pKa = 3.46DD681 pKa = 4.31LTTQFSWKK689 pKa = 9.53ILPPFPTRR697 pKa = 11.84EE698 pKa = 4.19EE699 pKa = 3.55IRR701 pKa = 11.84KK702 pKa = 9.61QISFYY707 pKa = 10.8HH708 pKa = 6.42GPLDD712 pKa = 4.94YY713 pKa = 10.97APKK716 pKa = 9.86WNFAHH721 pKa = 7.39FINKK725 pKa = 8.99PDD727 pKa = 4.0VIPPSSKK734 pKa = 9.47TMYY737 pKa = 10.16DD738 pKa = 3.47YY739 pKa = 10.95EE740 pKa = 4.68IEE742 pKa = 4.3HH743 pKa = 6.55SLRR746 pKa = 11.84PQPAPTFVARR756 pKa = 4.36
Molecular weight: 88.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0YTQ4|A0A0P0YTQ4_9VIRU RNA dependent RNA polymerase OS=Rosellinia necatrix partitivirus 6 OX=1573459 GN=RdRp PE=4 SV=1
MM1 pKa = 7.5SSKK4 pKa = 11.08LNLSDD9 pKa = 3.5RR10 pKa = 11.84AKK12 pKa = 10.46KK13 pKa = 10.41VSALKK18 pKa = 10.13TEE20 pKa = 4.37TTFKK24 pKa = 10.92AIDD27 pKa = 3.98LPGPTANFTTAFKK40 pKa = 10.59KK41 pKa = 10.86GSAPKK46 pKa = 9.38TGLANEE52 pKa = 4.53HH53 pKa = 5.86YY54 pKa = 10.72VYY56 pKa = 11.08VLPDD60 pKa = 3.79FSNILMFIIYY70 pKa = 9.47HH71 pKa = 7.15AIQFGPSLEE80 pKa = 4.37IKK82 pKa = 9.82NHH84 pKa = 5.93PKK86 pKa = 9.97VSIPVFVSYY95 pKa = 11.07CLTLVYY101 pKa = 11.04AHH103 pKa = 6.64FLVSDD108 pKa = 4.13AYY110 pKa = 9.83INPGQPHH117 pKa = 6.4FAMTIKK123 pKa = 10.61NDD125 pKa = 3.63SAYY128 pKa = 10.84SEE130 pKa = 4.33YY131 pKa = 11.21LDD133 pKa = 4.62FLLTLPVPEE142 pKa = 4.65FLTPLFAKK150 pKa = 8.2FTSTTTSRR158 pKa = 11.84RR159 pKa = 11.84TNTWFLASAEE169 pKa = 4.08GFMHH173 pKa = 6.2FTHH176 pKa = 7.45FGRR179 pKa = 11.84FFPINLFLNMHH190 pKa = 6.91DD191 pKa = 3.4LAARR195 pKa = 11.84TEE197 pKa = 4.33SRR199 pKa = 11.84ANPAEE204 pKa = 3.98VFYY207 pKa = 10.58DD208 pKa = 3.74LNMTEE213 pKa = 4.46IFHH216 pKa = 5.28ITKK219 pKa = 10.15YY220 pKa = 10.14RR221 pKa = 11.84DD222 pKa = 3.25PSVTTDD228 pKa = 2.98TAVTYY233 pKa = 10.05RR234 pKa = 11.84YY235 pKa = 10.12HH236 pKa = 7.13NFFASLYY243 pKa = 6.6PTTATTKK250 pKa = 8.87TRR252 pKa = 11.84RR253 pKa = 11.84SMNFFYY259 pKa = 10.44EE260 pKa = 4.05SRR262 pKa = 11.84INQVLEE268 pKa = 3.98GLFNPVLLRR277 pKa = 11.84AQQQRR282 pKa = 11.84QAFSRR287 pKa = 11.84ISIEE291 pKa = 3.84APTFEE296 pKa = 4.18TANYY300 pKa = 9.5NPYY303 pKa = 10.06FALLAITKK311 pKa = 9.54EE312 pKa = 4.15NIPEE316 pKa = 4.38LMTVMQSVADD326 pKa = 4.65TINGSIPIKK335 pKa = 10.27SQLNKK340 pKa = 10.1MYY342 pKa = 10.56DD343 pKa = 3.3NYY345 pKa = 11.25SGVSILDD352 pKa = 3.06HH353 pKa = 6.86GYY355 pKa = 11.23AGFPTPTWDD364 pKa = 4.3ADD366 pKa = 4.02SNLPDD371 pKa = 4.77NDD373 pKa = 3.46TTLDD377 pKa = 3.37KK378 pKa = 10.74FVYY381 pKa = 10.26NNAVLKK387 pKa = 10.85FEE389 pKa = 5.37GIPLSNYY396 pKa = 9.28AAAIKK401 pKa = 10.25FKK403 pKa = 10.89GAISSQSWTLSNNIIARR420 pKa = 11.84IDD422 pKa = 3.32RR423 pKa = 11.84SEE425 pKa = 4.15AVPNVTDD432 pKa = 4.15PAPDD436 pKa = 3.6TVIVTTTITPGSAYY450 pKa = 10.68LSLLALATVTDD461 pKa = 3.7RR462 pKa = 11.84HH463 pKa = 6.2DD464 pKa = 3.61VASSSTFYY472 pKa = 11.14LPNTASGPTDD482 pKa = 3.72EE483 pKa = 5.82NDD485 pKa = 3.62HH486 pKa = 5.35QHH488 pKa = 5.41MTTGNHH494 pKa = 5.19VLRR497 pKa = 11.84SYY499 pKa = 10.82SAEE502 pKa = 3.74LDD504 pKa = 3.29NYY506 pKa = 8.39PTVKK510 pKa = 10.22VLQFAEE516 pKa = 4.35TDD518 pKa = 3.74DD519 pKa = 3.68EE520 pKa = 4.43TAYY523 pKa = 10.78QATLFGTVIYY533 pKa = 10.49SDD535 pKa = 4.09EE536 pKa = 4.22LVGSIVNHH544 pKa = 5.69PHH546 pKa = 5.1PTSRR550 pKa = 11.84VEE552 pKa = 3.98EE553 pKa = 4.48DD554 pKa = 2.82NSLFLTSCIQFRR566 pKa = 11.84TTHH569 pKa = 6.79LATNFKK575 pKa = 9.45TNNPIASIIMKK586 pKa = 9.17TPAPTNDD593 pKa = 3.08AQFALSLYY601 pKa = 9.74RR602 pKa = 11.84SPNVTIPRR610 pKa = 11.84ITTHH614 pKa = 5.81TAHH617 pKa = 6.5SGYY620 pKa = 10.5HH621 pKa = 6.1RR622 pKa = 11.84DD623 pKa = 3.05TYY625 pKa = 11.12GLHH628 pKa = 7.13RR629 pKa = 11.84MTTNISVPEE638 pKa = 4.13FSQSILSYY646 pKa = 7.73TVRR649 pKa = 11.84DD650 pKa = 3.73RR651 pKa = 11.84PSRR654 pKa = 11.84RR655 pKa = 11.84HH656 pKa = 4.81QAPIHH661 pKa = 5.97EE662 pKa = 5.09PPSTPYY668 pKa = 10.61YY669 pKa = 10.57RR670 pKa = 11.84LLLWSPYY677 pKa = 10.6VFVTPNYY684 pKa = 9.9NRR686 pKa = 11.84DD687 pKa = 3.11WEE689 pKa = 4.1QDD691 pKa = 3.21EE692 pKa = 4.78FGQALLDD699 pKa = 3.45HH700 pKa = 7.03HH701 pKa = 7.11YY702 pKa = 9.33VTNLRR707 pKa = 11.84TIFGLDD713 pKa = 2.69ATMIEE718 pKa = 4.4VRR720 pKa = 11.84SGSAAMPVAA729 pKa = 4.54
MM1 pKa = 7.5SSKK4 pKa = 11.08LNLSDD9 pKa = 3.5RR10 pKa = 11.84AKK12 pKa = 10.46KK13 pKa = 10.41VSALKK18 pKa = 10.13TEE20 pKa = 4.37TTFKK24 pKa = 10.92AIDD27 pKa = 3.98LPGPTANFTTAFKK40 pKa = 10.59KK41 pKa = 10.86GSAPKK46 pKa = 9.38TGLANEE52 pKa = 4.53HH53 pKa = 5.86YY54 pKa = 10.72VYY56 pKa = 11.08VLPDD60 pKa = 3.79FSNILMFIIYY70 pKa = 9.47HH71 pKa = 7.15AIQFGPSLEE80 pKa = 4.37IKK82 pKa = 9.82NHH84 pKa = 5.93PKK86 pKa = 9.97VSIPVFVSYY95 pKa = 11.07CLTLVYY101 pKa = 11.04AHH103 pKa = 6.64FLVSDD108 pKa = 4.13AYY110 pKa = 9.83INPGQPHH117 pKa = 6.4FAMTIKK123 pKa = 10.61NDD125 pKa = 3.63SAYY128 pKa = 10.84SEE130 pKa = 4.33YY131 pKa = 11.21LDD133 pKa = 4.62FLLTLPVPEE142 pKa = 4.65FLTPLFAKK150 pKa = 8.2FTSTTTSRR158 pKa = 11.84RR159 pKa = 11.84TNTWFLASAEE169 pKa = 4.08GFMHH173 pKa = 6.2FTHH176 pKa = 7.45FGRR179 pKa = 11.84FFPINLFLNMHH190 pKa = 6.91DD191 pKa = 3.4LAARR195 pKa = 11.84TEE197 pKa = 4.33SRR199 pKa = 11.84ANPAEE204 pKa = 3.98VFYY207 pKa = 10.58DD208 pKa = 3.74LNMTEE213 pKa = 4.46IFHH216 pKa = 5.28ITKK219 pKa = 10.15YY220 pKa = 10.14RR221 pKa = 11.84DD222 pKa = 3.25PSVTTDD228 pKa = 2.98TAVTYY233 pKa = 10.05RR234 pKa = 11.84YY235 pKa = 10.12HH236 pKa = 7.13NFFASLYY243 pKa = 6.6PTTATTKK250 pKa = 8.87TRR252 pKa = 11.84RR253 pKa = 11.84SMNFFYY259 pKa = 10.44EE260 pKa = 4.05SRR262 pKa = 11.84INQVLEE268 pKa = 3.98GLFNPVLLRR277 pKa = 11.84AQQQRR282 pKa = 11.84QAFSRR287 pKa = 11.84ISIEE291 pKa = 3.84APTFEE296 pKa = 4.18TANYY300 pKa = 9.5NPYY303 pKa = 10.06FALLAITKK311 pKa = 9.54EE312 pKa = 4.15NIPEE316 pKa = 4.38LMTVMQSVADD326 pKa = 4.65TINGSIPIKK335 pKa = 10.27SQLNKK340 pKa = 10.1MYY342 pKa = 10.56DD343 pKa = 3.3NYY345 pKa = 11.25SGVSILDD352 pKa = 3.06HH353 pKa = 6.86GYY355 pKa = 11.23AGFPTPTWDD364 pKa = 4.3ADD366 pKa = 4.02SNLPDD371 pKa = 4.77NDD373 pKa = 3.46TTLDD377 pKa = 3.37KK378 pKa = 10.74FVYY381 pKa = 10.26NNAVLKK387 pKa = 10.85FEE389 pKa = 5.37GIPLSNYY396 pKa = 9.28AAAIKK401 pKa = 10.25FKK403 pKa = 10.89GAISSQSWTLSNNIIARR420 pKa = 11.84IDD422 pKa = 3.32RR423 pKa = 11.84SEE425 pKa = 4.15AVPNVTDD432 pKa = 4.15PAPDD436 pKa = 3.6TVIVTTTITPGSAYY450 pKa = 10.68LSLLALATVTDD461 pKa = 3.7RR462 pKa = 11.84HH463 pKa = 6.2DD464 pKa = 3.61VASSSTFYY472 pKa = 11.14LPNTASGPTDD482 pKa = 3.72EE483 pKa = 5.82NDD485 pKa = 3.62HH486 pKa = 5.35QHH488 pKa = 5.41MTTGNHH494 pKa = 5.19VLRR497 pKa = 11.84SYY499 pKa = 10.82SAEE502 pKa = 3.74LDD504 pKa = 3.29NYY506 pKa = 8.39PTVKK510 pKa = 10.22VLQFAEE516 pKa = 4.35TDD518 pKa = 3.74DD519 pKa = 3.68EE520 pKa = 4.43TAYY523 pKa = 10.78QATLFGTVIYY533 pKa = 10.49SDD535 pKa = 4.09EE536 pKa = 4.22LVGSIVNHH544 pKa = 5.69PHH546 pKa = 5.1PTSRR550 pKa = 11.84VEE552 pKa = 3.98EE553 pKa = 4.48DD554 pKa = 2.82NSLFLTSCIQFRR566 pKa = 11.84TTHH569 pKa = 6.79LATNFKK575 pKa = 9.45TNNPIASIIMKK586 pKa = 9.17TPAPTNDD593 pKa = 3.08AQFALSLYY601 pKa = 9.74RR602 pKa = 11.84SPNVTIPRR610 pKa = 11.84ITTHH614 pKa = 5.81TAHH617 pKa = 6.5SGYY620 pKa = 10.5HH621 pKa = 6.1RR622 pKa = 11.84DD623 pKa = 3.05TYY625 pKa = 11.12GLHH628 pKa = 7.13RR629 pKa = 11.84MTTNISVPEE638 pKa = 4.13FSQSILSYY646 pKa = 7.73TVRR649 pKa = 11.84DD650 pKa = 3.73RR651 pKa = 11.84PSRR654 pKa = 11.84RR655 pKa = 11.84HH656 pKa = 4.81QAPIHH661 pKa = 5.97EE662 pKa = 5.09PPSTPYY668 pKa = 10.61YY669 pKa = 10.57RR670 pKa = 11.84LLLWSPYY677 pKa = 10.6VFVTPNYY684 pKa = 9.9NRR686 pKa = 11.84DD687 pKa = 3.11WEE689 pKa = 4.1QDD691 pKa = 3.21EE692 pKa = 4.78FGQALLDD699 pKa = 3.45HH700 pKa = 7.03HH701 pKa = 7.11YY702 pKa = 9.33VTNLRR707 pKa = 11.84TIFGLDD713 pKa = 2.69ATMIEE718 pKa = 4.4VRR720 pKa = 11.84SGSAAMPVAA729 pKa = 4.54
Molecular weight: 81.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1485 |
729 |
756 |
742.5 |
85.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.936 ± 0.89 | 0.741 ± 0.321 |
6.128 ± 0.724 | 4.579 ± 0.413 |
5.993 ± 0.123 | 3.704 ± 0.283 |
3.569 ± 0.002 | 6.061 ± 0.206 |
3.838 ± 0.376 | 8.485 ± 0.081 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.155 ± 0.067 | 5.32 ± 0.587 |
6.465 ± 0.082 | 3.502 ± 0.616 |
5.387 ± 0.686 | 6.936 ± 0.89 |
9.091 ± 1.2 | 4.31 ± 0.809 |
0.808 ± 0.084 | 5.993 ± 0.725 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
