
Microbacterium phage ChickenKing
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Schubertvirus; unclassified Schubertvirus
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
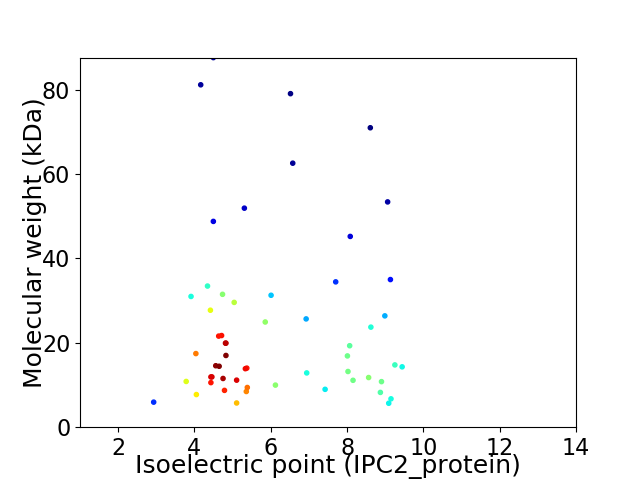
Virtual 2D-PAGE plot for 57 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
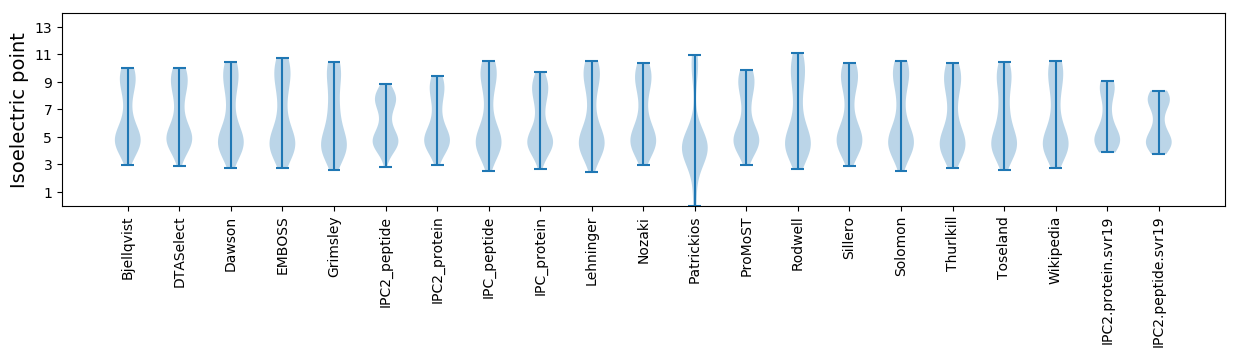
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J6T8S3|A0A5J6T8S3_9CAUD Scaffolding protein OS=Microbacterium phage ChickenKing OX=2652406 GN=5 PE=4 SV=1
MM1 pKa = 7.51EE2 pKa = 5.12NNYY5 pKa = 8.3PTFQEE10 pKa = 3.85VRR12 pKa = 11.84NYY14 pKa = 10.72LINDD18 pKa = 3.81NDD20 pKa = 3.48WTAEE24 pKa = 4.04EE25 pKa = 4.27VDD27 pKa = 3.72QLAGYY32 pKa = 9.83AFRR35 pKa = 11.84HH36 pKa = 5.65INQDD40 pKa = 2.44GTTTMTRR47 pKa = 11.84DD48 pKa = 2.52QWTIYY53 pKa = 10.34IMDD56 pKa = 4.08YY57 pKa = 10.82CNEE60 pKa = 4.55HH61 pKa = 6.81IDD63 pKa = 3.8LL64 pKa = 4.85
MM1 pKa = 7.51EE2 pKa = 5.12NNYY5 pKa = 8.3PTFQEE10 pKa = 3.85VRR12 pKa = 11.84NYY14 pKa = 10.72LINDD18 pKa = 3.81NDD20 pKa = 3.48WTAEE24 pKa = 4.04EE25 pKa = 4.27VDD27 pKa = 3.72QLAGYY32 pKa = 9.83AFRR35 pKa = 11.84HH36 pKa = 5.65INQDD40 pKa = 2.44GTTTMTRR47 pKa = 11.84DD48 pKa = 2.52QWTIYY53 pKa = 10.34IMDD56 pKa = 4.08YY57 pKa = 10.82CNEE60 pKa = 4.55HH61 pKa = 6.81IDD63 pKa = 3.8LL64 pKa = 4.85
Molecular weight: 7.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J6T5S0|A0A5J6T5S0_9CAUD Uncharacterized protein OS=Microbacterium phage ChickenKing OX=2652406 GN=50 PE=4 SV=1
MM1 pKa = 7.18TLVLRR6 pKa = 11.84DD7 pKa = 3.77AQRR10 pKa = 11.84VALEE14 pKa = 4.05RR15 pKa = 11.84LTEE18 pKa = 4.35PGRR21 pKa = 11.84HH22 pKa = 5.61FAALWAEE29 pKa = 4.02PRR31 pKa = 11.84SGKK34 pKa = 8.81TGVALSWIQHH44 pKa = 4.36VKK46 pKa = 9.75PRR48 pKa = 11.84VAVVVGPKK56 pKa = 9.68VAEE59 pKa = 4.0ATWRR63 pKa = 11.84SEE65 pKa = 3.64AAKK68 pKa = 9.28WLTMEE73 pKa = 4.08YY74 pKa = 10.5RR75 pKa = 11.84FFPLTTGNEE84 pKa = 4.26YY85 pKa = 10.33PDD87 pKa = 3.4TRR89 pKa = 11.84SFKK92 pKa = 9.77GTTILFVNYY101 pKa = 10.0DD102 pKa = 3.82QFAKK106 pKa = 10.78APWKK110 pKa = 10.55RR111 pKa = 11.84LGPFLKK117 pKa = 10.54KK118 pKa = 9.94MSEE121 pKa = 4.28YY122 pKa = 10.74CQGQGMMLLDD132 pKa = 4.07EE133 pKa = 4.33SHH135 pKa = 7.05IIKK138 pKa = 9.26TPSSVIGRR146 pKa = 11.84NIRR149 pKa = 11.84PLADD153 pKa = 2.73KK154 pKa = 9.38WHH156 pKa = 6.15YY157 pKa = 9.9RR158 pKa = 11.84LLMTGTPVTNPNQIDD173 pKa = 3.49AVYY176 pKa = 9.75GQWTFLDD183 pKa = 4.07PSIRR187 pKa = 11.84DD188 pKa = 3.45HH189 pKa = 6.51WPSARR194 pKa = 11.84DD195 pKa = 3.26FRR197 pKa = 11.84EE198 pKa = 4.06YY199 pKa = 10.4FGEE202 pKa = 3.92WTNVKK207 pKa = 10.04GFPEE211 pKa = 5.13LIKK214 pKa = 10.3PRR216 pKa = 11.84HH217 pKa = 4.5QAEE220 pKa = 4.21LSLYY224 pKa = 8.41LQPNVITMVGPNDD237 pKa = 3.63PVPIRR242 pKa = 11.84KK243 pKa = 8.51VRR245 pKa = 11.84YY246 pKa = 7.52PVPEE250 pKa = 4.54EE251 pKa = 3.47IRR253 pKa = 11.84LQLKK257 pKa = 9.51EE258 pKa = 4.05LFKK261 pKa = 11.07EE262 pKa = 4.74GVVEE266 pKa = 4.29VQGHH270 pKa = 5.97TIAALNPLTRR280 pKa = 11.84LLRR283 pKa = 11.84MRR285 pKa = 11.84SLVAGWAKK293 pKa = 10.88DD294 pKa = 3.67DD295 pKa = 4.09EE296 pKa = 4.92GTSFTVPEE304 pKa = 3.99AARR307 pKa = 11.84ARR309 pKa = 11.84LRR311 pKa = 11.84ALSHH315 pKa = 5.59IVKK318 pKa = 10.04KK319 pKa = 9.61RR320 pKa = 11.84TEE322 pKa = 4.03GKK324 pKa = 10.51VIIACTHH331 pKa = 5.21LWEE334 pKa = 4.19VRR336 pKa = 11.84LVKK339 pKa = 10.41GWMRR343 pKa = 11.84RR344 pKa = 11.84QGIGYY349 pKa = 9.56RR350 pKa = 11.84IITGDD355 pKa = 3.52TKK357 pKa = 11.02EE358 pKa = 4.16KK359 pKa = 10.42NHH361 pKa = 6.58VIEE364 pKa = 4.96DD365 pKa = 3.72FQYY368 pKa = 11.46DD369 pKa = 3.77RR370 pKa = 11.84DD371 pKa = 4.26CKK373 pKa = 10.92VLLVQPRR380 pKa = 11.84TVAMAVDD387 pKa = 3.63ISVANDD393 pKa = 4.38LIWYY397 pKa = 7.42TSDD400 pKa = 3.05FNYY403 pKa = 9.15VTFKK407 pKa = 10.53QASDD411 pKa = 3.99RR412 pKa = 11.84IKK414 pKa = 10.76LSPASPTVWFLCGKK428 pKa = 8.52GTVDD432 pKa = 3.16EE433 pKa = 5.37DD434 pKa = 3.51VWKK437 pKa = 8.91TLQEE441 pKa = 4.24DD442 pKa = 3.71HH443 pKa = 7.37DD444 pKa = 4.54HH445 pKa = 6.72LNKK448 pKa = 9.69TVAKK452 pKa = 9.87IKK454 pKa = 9.35RR455 pKa = 11.84QKK457 pKa = 8.36RR458 pKa = 11.84HH459 pKa = 5.43PYY461 pKa = 9.58RR462 pKa = 11.84RR463 pKa = 3.54
MM1 pKa = 7.18TLVLRR6 pKa = 11.84DD7 pKa = 3.77AQRR10 pKa = 11.84VALEE14 pKa = 4.05RR15 pKa = 11.84LTEE18 pKa = 4.35PGRR21 pKa = 11.84HH22 pKa = 5.61FAALWAEE29 pKa = 4.02PRR31 pKa = 11.84SGKK34 pKa = 8.81TGVALSWIQHH44 pKa = 4.36VKK46 pKa = 9.75PRR48 pKa = 11.84VAVVVGPKK56 pKa = 9.68VAEE59 pKa = 4.0ATWRR63 pKa = 11.84SEE65 pKa = 3.64AAKK68 pKa = 9.28WLTMEE73 pKa = 4.08YY74 pKa = 10.5RR75 pKa = 11.84FFPLTTGNEE84 pKa = 4.26YY85 pKa = 10.33PDD87 pKa = 3.4TRR89 pKa = 11.84SFKK92 pKa = 9.77GTTILFVNYY101 pKa = 10.0DD102 pKa = 3.82QFAKK106 pKa = 10.78APWKK110 pKa = 10.55RR111 pKa = 11.84LGPFLKK117 pKa = 10.54KK118 pKa = 9.94MSEE121 pKa = 4.28YY122 pKa = 10.74CQGQGMMLLDD132 pKa = 4.07EE133 pKa = 4.33SHH135 pKa = 7.05IIKK138 pKa = 9.26TPSSVIGRR146 pKa = 11.84NIRR149 pKa = 11.84PLADD153 pKa = 2.73KK154 pKa = 9.38WHH156 pKa = 6.15YY157 pKa = 9.9RR158 pKa = 11.84LLMTGTPVTNPNQIDD173 pKa = 3.49AVYY176 pKa = 9.75GQWTFLDD183 pKa = 4.07PSIRR187 pKa = 11.84DD188 pKa = 3.45HH189 pKa = 6.51WPSARR194 pKa = 11.84DD195 pKa = 3.26FRR197 pKa = 11.84EE198 pKa = 4.06YY199 pKa = 10.4FGEE202 pKa = 3.92WTNVKK207 pKa = 10.04GFPEE211 pKa = 5.13LIKK214 pKa = 10.3PRR216 pKa = 11.84HH217 pKa = 4.5QAEE220 pKa = 4.21LSLYY224 pKa = 8.41LQPNVITMVGPNDD237 pKa = 3.63PVPIRR242 pKa = 11.84KK243 pKa = 8.51VRR245 pKa = 11.84YY246 pKa = 7.52PVPEE250 pKa = 4.54EE251 pKa = 3.47IRR253 pKa = 11.84LQLKK257 pKa = 9.51EE258 pKa = 4.05LFKK261 pKa = 11.07EE262 pKa = 4.74GVVEE266 pKa = 4.29VQGHH270 pKa = 5.97TIAALNPLTRR280 pKa = 11.84LLRR283 pKa = 11.84MRR285 pKa = 11.84SLVAGWAKK293 pKa = 10.88DD294 pKa = 3.67DD295 pKa = 4.09EE296 pKa = 4.92GTSFTVPEE304 pKa = 3.99AARR307 pKa = 11.84ARR309 pKa = 11.84LRR311 pKa = 11.84ALSHH315 pKa = 5.59IVKK318 pKa = 10.04KK319 pKa = 9.61RR320 pKa = 11.84TEE322 pKa = 4.03GKK324 pKa = 10.51VIIACTHH331 pKa = 5.21LWEE334 pKa = 4.19VRR336 pKa = 11.84LVKK339 pKa = 10.41GWMRR343 pKa = 11.84RR344 pKa = 11.84QGIGYY349 pKa = 9.56RR350 pKa = 11.84IITGDD355 pKa = 3.52TKK357 pKa = 11.02EE358 pKa = 4.16KK359 pKa = 10.42NHH361 pKa = 6.58VIEE364 pKa = 4.96DD365 pKa = 3.72FQYY368 pKa = 11.46DD369 pKa = 3.77RR370 pKa = 11.84DD371 pKa = 4.26CKK373 pKa = 10.92VLLVQPRR380 pKa = 11.84TVAMAVDD387 pKa = 3.63ISVANDD393 pKa = 4.38LIWYY397 pKa = 7.42TSDD400 pKa = 3.05FNYY403 pKa = 9.15VTFKK407 pKa = 10.53QASDD411 pKa = 3.99RR412 pKa = 11.84IKK414 pKa = 10.76LSPASPTVWFLCGKK428 pKa = 8.52GTVDD432 pKa = 3.16EE433 pKa = 5.37DD434 pKa = 3.51VWKK437 pKa = 8.91TLQEE441 pKa = 4.24DD442 pKa = 3.71HH443 pKa = 7.37DD444 pKa = 4.54HH445 pKa = 6.72LNKK448 pKa = 9.69TVAKK452 pKa = 9.87IKK454 pKa = 9.35RR455 pKa = 11.84QKK457 pKa = 8.36RR458 pKa = 11.84HH459 pKa = 5.43PYY461 pKa = 9.58RR462 pKa = 11.84RR463 pKa = 3.54
Molecular weight: 53.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12601 |
50 |
808 |
221.1 |
24.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.015 ± 0.424 | 0.651 ± 0.097 |
6.468 ± 0.307 | 6.484 ± 0.4 |
3.024 ± 0.226 | 8.039 ± 0.38 |
1.873 ± 0.196 | 4.857 ± 0.409 |
4.881 ± 0.398 | 8.46 ± 0.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.124 | 3.531 ± 0.185 |
4.539 ± 0.225 | 3.952 ± 0.167 |
5.754 ± 0.358 | 5.928 ± 0.327 |
6.706 ± 0.299 | 7.428 ± 0.32 |
2.016 ± 0.205 | 2.944 ± 0.221 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
