
Beihai sobemo-like virus 19
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.73
Get precalculated fractions of proteins
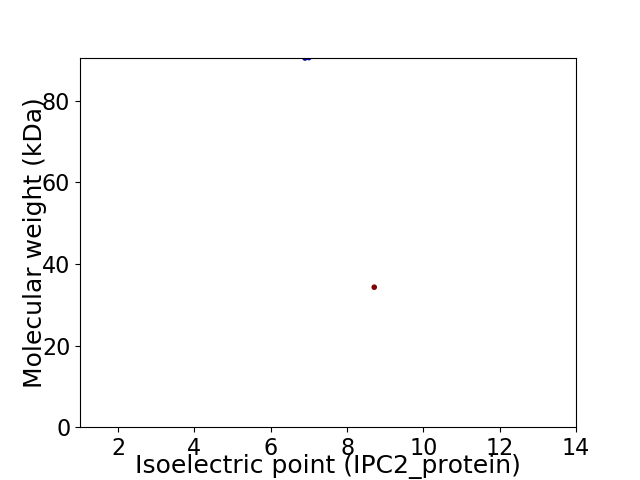
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
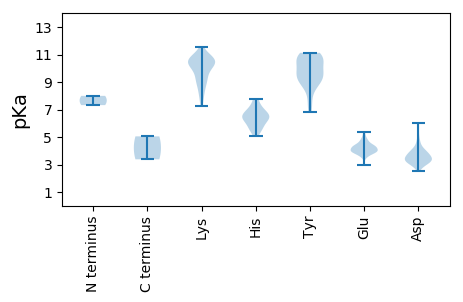
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE63|A0A1L3KE63_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 19 OX=1922690 PE=4 SV=1
MM1 pKa = 7.99AIPSFIASMYY11 pKa = 10.93DD12 pKa = 3.25LASEE16 pKa = 4.11QFGVIGMLKK25 pKa = 10.51PNLTLNWDD33 pKa = 3.9YY34 pKa = 10.8FQSKK38 pKa = 10.55APTSPPEE45 pKa = 3.85VQTFAGQLLASIMVSVYY62 pKa = 10.96SLYY65 pKa = 10.94KK66 pKa = 8.42STDD69 pKa = 3.62MIWAFLVGIVTSLNPNIRR87 pKa = 11.84TSNLVRR93 pKa = 11.84PFTIDD98 pKa = 4.03KK99 pKa = 9.25ILSYY103 pKa = 11.13AGLCTCAYY111 pKa = 9.04WSYY114 pKa = 9.24TAEE117 pKa = 4.05NVIMAFFAVVGMCQSAKK134 pKa = 10.59LVIGHH139 pKa = 7.23LWWCKK144 pKa = 8.96NAWSASEE151 pKa = 4.71HH152 pKa = 6.31SDD154 pKa = 3.38QIAGNSYY161 pKa = 9.89FEE163 pKa = 5.22SMTADD168 pKa = 3.28SSIKK172 pKa = 10.48EE173 pKa = 4.08DD174 pKa = 3.5TKK176 pKa = 11.58KK177 pKa = 10.22MKK179 pKa = 9.89CQAMACAGPYY189 pKa = 8.94TGSPLVGQVIRR200 pKa = 11.84LPSFMDD206 pKa = 4.74DD207 pKa = 3.34GEE209 pKa = 4.8LYY211 pKa = 7.89CTPVHH216 pKa = 6.24VLNDD220 pKa = 4.07DD221 pKa = 3.39VTHH224 pKa = 6.63IVTHH228 pKa = 6.36KK229 pKa = 10.44SSVNIEE235 pKa = 4.31SVEE238 pKa = 3.95PLPPARR244 pKa = 11.84GFPDD248 pKa = 3.7LAFLKK253 pKa = 10.59LPNGTGAQLGMSEE266 pKa = 4.59AVVSSIMGGEE276 pKa = 3.91TGMAHH281 pKa = 6.85AMGVGSNFYY290 pKa = 9.63EE291 pKa = 4.29RR292 pKa = 11.84SSGRR296 pKa = 11.84IIEE299 pKa = 4.15LEE301 pKa = 4.02RR302 pKa = 11.84GGLAYY307 pKa = 10.59LGSTLPGWSGAPYY320 pKa = 9.57EE321 pKa = 4.29VGGKK325 pKa = 9.39VVGVHH330 pKa = 5.65HH331 pKa = 6.46VGIPSEE337 pKa = 4.29GNLGYY342 pKa = 10.72DD343 pKa = 3.23ILLAMKK349 pKa = 10.07ILEE352 pKa = 4.29KK353 pKa = 10.72QMQEE357 pKa = 3.54PEE359 pKa = 3.89YY360 pKa = 10.22EE361 pKa = 3.91KK362 pKa = 10.7RR363 pKa = 11.84KK364 pKa = 10.4RR365 pKa = 11.84GWKK368 pKa = 9.58KK369 pKa = 9.01KK370 pKa = 8.75FRR372 pKa = 11.84RR373 pKa = 11.84EE374 pKa = 3.9KK375 pKa = 11.06YY376 pKa = 8.49PVADD380 pKa = 3.2IRR382 pKa = 11.84ASNGYY387 pKa = 9.83AISEE391 pKa = 4.07HH392 pKa = 6.82RR393 pKa = 11.84IGGARR398 pKa = 11.84TDD400 pKa = 3.52IMWKK404 pKa = 9.72IKK406 pKa = 10.19NPKK409 pKa = 9.06GKK411 pKa = 9.55IVMLYY416 pKa = 10.4YY417 pKa = 10.71DD418 pKa = 5.03DD419 pKa = 3.98LTEE422 pKa = 4.23EE423 pKa = 4.35EE424 pKa = 4.39RR425 pKa = 11.84SHH427 pKa = 7.78IRR429 pKa = 11.84GEE431 pKa = 4.11MDD433 pKa = 2.56WQGYY437 pKa = 7.32SFEE440 pKa = 4.98SGPKK444 pKa = 8.17PTTPVNAGLIEE455 pKa = 3.96QVRR458 pKa = 11.84NNLRR462 pKa = 11.84AAQNAKK468 pKa = 10.08RR469 pKa = 11.84DD470 pKa = 3.96LQTMSDD476 pKa = 4.46LYY478 pKa = 11.06QLANIRR484 pKa = 11.84TRR486 pKa = 11.84NGPLEE491 pKa = 3.9TAAQVGVDD499 pKa = 3.67TQAINAAISEE509 pKa = 4.32LHH511 pKa = 5.89LQMEE515 pKa = 5.03RR516 pKa = 11.84FAEE519 pKa = 4.67LIEE522 pKa = 4.31YY523 pKa = 10.28YY524 pKa = 9.74SAKK527 pKa = 10.04SAEE530 pKa = 4.82DD531 pKa = 3.18IQNQKK536 pKa = 10.48RR537 pKa = 11.84MIRR540 pKa = 11.84KK541 pKa = 8.82KK542 pKa = 10.35RR543 pKa = 11.84NKK545 pKa = 9.45EE546 pKa = 3.77KK547 pKa = 10.45PLVSQEE553 pKa = 3.76VAAEE557 pKa = 4.07IQRR560 pKa = 11.84KK561 pKa = 7.49QADD564 pKa = 3.16INAALSRR571 pKa = 11.84AAHH574 pKa = 6.27LPPRR578 pKa = 11.84EE579 pKa = 3.92QGGQIDD585 pKa = 4.55DD586 pKa = 4.27EE587 pKa = 5.35NDD589 pKa = 3.13VEE591 pKa = 4.39MPAYY595 pKa = 9.24EE596 pKa = 4.26KK597 pKa = 10.86LSSHH601 pKa = 5.92FVGRR605 pKa = 11.84FKK607 pKa = 11.06DD608 pKa = 3.28KK609 pKa = 10.71YY610 pKa = 9.49GAIPRR615 pKa = 11.84EE616 pKa = 3.92FLDD619 pKa = 4.07DD620 pKa = 3.9CSSGEE625 pKa = 3.96EE626 pKa = 4.12SGEE629 pKa = 4.21EE630 pKa = 3.77EE631 pKa = 4.82DD632 pKa = 5.55SPTFSPTPSMEE643 pKa = 3.96KK644 pKa = 10.27SSLNVDD650 pKa = 3.04QDD652 pKa = 3.72ASSTSCQTTKK662 pKa = 10.8TDD664 pKa = 3.56VEE666 pKa = 4.4RR667 pKa = 11.84LKK669 pKa = 10.82HH670 pKa = 6.48LYY672 pKa = 8.96MSKK675 pKa = 9.92LHH677 pKa = 6.21SGQMEE682 pKa = 4.38ATSGPQMINQTSLNRR697 pKa = 11.84LKK699 pKa = 10.22NTFNEE704 pKa = 4.18DD705 pKa = 2.86MRR707 pKa = 11.84QLEE710 pKa = 4.59VASLPMTPKK719 pKa = 10.71LGRR722 pKa = 11.84LQSAFDD728 pKa = 3.66TSLGGSKK735 pKa = 8.07TQFNYY740 pKa = 9.87EE741 pKa = 3.61QSGKK745 pKa = 10.21KK746 pKa = 9.29KK747 pKa = 10.03RR748 pKa = 11.84KK749 pKa = 7.64SVHH752 pKa = 5.11WKK754 pKa = 8.1TSKK757 pKa = 10.26KK758 pKa = 10.25RR759 pKa = 11.84NSQTSSKK766 pKa = 10.59RR767 pKa = 11.84SLTRR771 pKa = 11.84NPVQEE776 pKa = 4.37SHH778 pKa = 6.68TSTKK782 pKa = 10.37DD783 pKa = 3.12KK784 pKa = 11.13QMQKK788 pKa = 10.37SLDD791 pKa = 3.61MSSSKK796 pKa = 9.53DD797 pKa = 3.44TQMSMASDD805 pKa = 3.89RR806 pKa = 11.84MQSTEE811 pKa = 3.83SGNN814 pKa = 3.38
MM1 pKa = 7.99AIPSFIASMYY11 pKa = 10.93DD12 pKa = 3.25LASEE16 pKa = 4.11QFGVIGMLKK25 pKa = 10.51PNLTLNWDD33 pKa = 3.9YY34 pKa = 10.8FQSKK38 pKa = 10.55APTSPPEE45 pKa = 3.85VQTFAGQLLASIMVSVYY62 pKa = 10.96SLYY65 pKa = 10.94KK66 pKa = 8.42STDD69 pKa = 3.62MIWAFLVGIVTSLNPNIRR87 pKa = 11.84TSNLVRR93 pKa = 11.84PFTIDD98 pKa = 4.03KK99 pKa = 9.25ILSYY103 pKa = 11.13AGLCTCAYY111 pKa = 9.04WSYY114 pKa = 9.24TAEE117 pKa = 4.05NVIMAFFAVVGMCQSAKK134 pKa = 10.59LVIGHH139 pKa = 7.23LWWCKK144 pKa = 8.96NAWSASEE151 pKa = 4.71HH152 pKa = 6.31SDD154 pKa = 3.38QIAGNSYY161 pKa = 9.89FEE163 pKa = 5.22SMTADD168 pKa = 3.28SSIKK172 pKa = 10.48EE173 pKa = 4.08DD174 pKa = 3.5TKK176 pKa = 11.58KK177 pKa = 10.22MKK179 pKa = 9.89CQAMACAGPYY189 pKa = 8.94TGSPLVGQVIRR200 pKa = 11.84LPSFMDD206 pKa = 4.74DD207 pKa = 3.34GEE209 pKa = 4.8LYY211 pKa = 7.89CTPVHH216 pKa = 6.24VLNDD220 pKa = 4.07DD221 pKa = 3.39VTHH224 pKa = 6.63IVTHH228 pKa = 6.36KK229 pKa = 10.44SSVNIEE235 pKa = 4.31SVEE238 pKa = 3.95PLPPARR244 pKa = 11.84GFPDD248 pKa = 3.7LAFLKK253 pKa = 10.59LPNGTGAQLGMSEE266 pKa = 4.59AVVSSIMGGEE276 pKa = 3.91TGMAHH281 pKa = 6.85AMGVGSNFYY290 pKa = 9.63EE291 pKa = 4.29RR292 pKa = 11.84SSGRR296 pKa = 11.84IIEE299 pKa = 4.15LEE301 pKa = 4.02RR302 pKa = 11.84GGLAYY307 pKa = 10.59LGSTLPGWSGAPYY320 pKa = 9.57EE321 pKa = 4.29VGGKK325 pKa = 9.39VVGVHH330 pKa = 5.65HH331 pKa = 6.46VGIPSEE337 pKa = 4.29GNLGYY342 pKa = 10.72DD343 pKa = 3.23ILLAMKK349 pKa = 10.07ILEE352 pKa = 4.29KK353 pKa = 10.72QMQEE357 pKa = 3.54PEE359 pKa = 3.89YY360 pKa = 10.22EE361 pKa = 3.91KK362 pKa = 10.7RR363 pKa = 11.84KK364 pKa = 10.4RR365 pKa = 11.84GWKK368 pKa = 9.58KK369 pKa = 9.01KK370 pKa = 8.75FRR372 pKa = 11.84RR373 pKa = 11.84EE374 pKa = 3.9KK375 pKa = 11.06YY376 pKa = 8.49PVADD380 pKa = 3.2IRR382 pKa = 11.84ASNGYY387 pKa = 9.83AISEE391 pKa = 4.07HH392 pKa = 6.82RR393 pKa = 11.84IGGARR398 pKa = 11.84TDD400 pKa = 3.52IMWKK404 pKa = 9.72IKK406 pKa = 10.19NPKK409 pKa = 9.06GKK411 pKa = 9.55IVMLYY416 pKa = 10.4YY417 pKa = 10.71DD418 pKa = 5.03DD419 pKa = 3.98LTEE422 pKa = 4.23EE423 pKa = 4.35EE424 pKa = 4.39RR425 pKa = 11.84SHH427 pKa = 7.78IRR429 pKa = 11.84GEE431 pKa = 4.11MDD433 pKa = 2.56WQGYY437 pKa = 7.32SFEE440 pKa = 4.98SGPKK444 pKa = 8.17PTTPVNAGLIEE455 pKa = 3.96QVRR458 pKa = 11.84NNLRR462 pKa = 11.84AAQNAKK468 pKa = 10.08RR469 pKa = 11.84DD470 pKa = 3.96LQTMSDD476 pKa = 4.46LYY478 pKa = 11.06QLANIRR484 pKa = 11.84TRR486 pKa = 11.84NGPLEE491 pKa = 3.9TAAQVGVDD499 pKa = 3.67TQAINAAISEE509 pKa = 4.32LHH511 pKa = 5.89LQMEE515 pKa = 5.03RR516 pKa = 11.84FAEE519 pKa = 4.67LIEE522 pKa = 4.31YY523 pKa = 10.28YY524 pKa = 9.74SAKK527 pKa = 10.04SAEE530 pKa = 4.82DD531 pKa = 3.18IQNQKK536 pKa = 10.48RR537 pKa = 11.84MIRR540 pKa = 11.84KK541 pKa = 8.82KK542 pKa = 10.35RR543 pKa = 11.84NKK545 pKa = 9.45EE546 pKa = 3.77KK547 pKa = 10.45PLVSQEE553 pKa = 3.76VAAEE557 pKa = 4.07IQRR560 pKa = 11.84KK561 pKa = 7.49QADD564 pKa = 3.16INAALSRR571 pKa = 11.84AAHH574 pKa = 6.27LPPRR578 pKa = 11.84EE579 pKa = 3.92QGGQIDD585 pKa = 4.55DD586 pKa = 4.27EE587 pKa = 5.35NDD589 pKa = 3.13VEE591 pKa = 4.39MPAYY595 pKa = 9.24EE596 pKa = 4.26KK597 pKa = 10.86LSSHH601 pKa = 5.92FVGRR605 pKa = 11.84FKK607 pKa = 11.06DD608 pKa = 3.28KK609 pKa = 10.71YY610 pKa = 9.49GAIPRR615 pKa = 11.84EE616 pKa = 3.92FLDD619 pKa = 4.07DD620 pKa = 3.9CSSGEE625 pKa = 3.96EE626 pKa = 4.12SGEE629 pKa = 4.21EE630 pKa = 3.77EE631 pKa = 4.82DD632 pKa = 5.55SPTFSPTPSMEE643 pKa = 3.96KK644 pKa = 10.27SSLNVDD650 pKa = 3.04QDD652 pKa = 3.72ASSTSCQTTKK662 pKa = 10.8TDD664 pKa = 3.56VEE666 pKa = 4.4RR667 pKa = 11.84LKK669 pKa = 10.82HH670 pKa = 6.48LYY672 pKa = 8.96MSKK675 pKa = 9.92LHH677 pKa = 6.21SGQMEE682 pKa = 4.38ATSGPQMINQTSLNRR697 pKa = 11.84LKK699 pKa = 10.22NTFNEE704 pKa = 4.18DD705 pKa = 2.86MRR707 pKa = 11.84QLEE710 pKa = 4.59VASLPMTPKK719 pKa = 10.71LGRR722 pKa = 11.84LQSAFDD728 pKa = 3.66TSLGGSKK735 pKa = 8.07TQFNYY740 pKa = 9.87EE741 pKa = 3.61QSGKK745 pKa = 10.21KK746 pKa = 9.29KK747 pKa = 10.03RR748 pKa = 11.84KK749 pKa = 7.64SVHH752 pKa = 5.11WKK754 pKa = 8.1TSKK757 pKa = 10.26KK758 pKa = 10.25RR759 pKa = 11.84NSQTSSKK766 pKa = 10.59RR767 pKa = 11.84SLTRR771 pKa = 11.84NPVQEE776 pKa = 4.37SHH778 pKa = 6.68TSTKK782 pKa = 10.37DD783 pKa = 3.12KK784 pKa = 11.13QMQKK788 pKa = 10.37SLDD791 pKa = 3.61MSSSKK796 pKa = 9.53DD797 pKa = 3.44TQMSMASDD805 pKa = 3.89RR806 pKa = 11.84MQSTEE811 pKa = 3.83SGNN814 pKa = 3.38
Molecular weight: 90.48 kDa
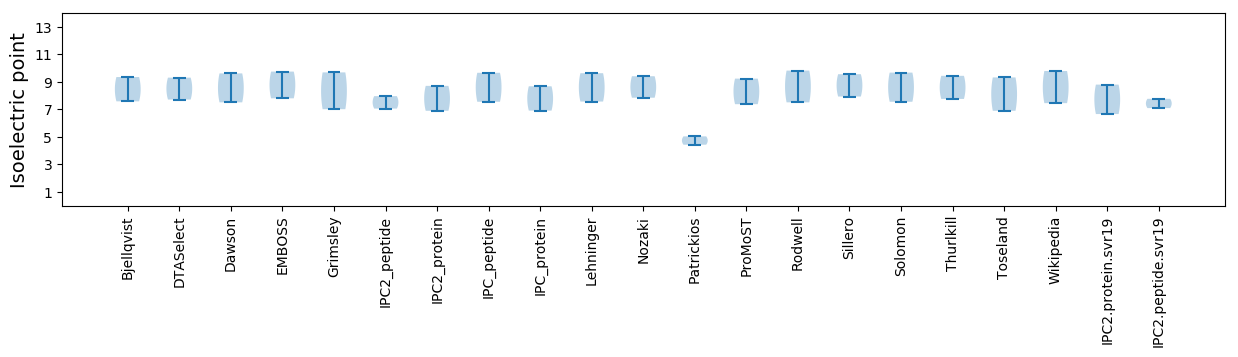
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE63|A0A1L3KE63_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 19 OX=1922690 PE=4 SV=1
MM1 pKa = 7.32WRR3 pKa = 11.84LISGVSVRR11 pKa = 11.84DD12 pKa = 3.34QVVAHH17 pKa = 5.81YY18 pKa = 10.74LFGDD22 pKa = 3.47MMEE25 pKa = 4.47VLIEE29 pKa = 4.14HH30 pKa = 7.28PGLYY34 pKa = 8.59HH35 pKa = 7.12TMIGWSPFQRR45 pKa = 11.84AGQRR49 pKa = 11.84YY50 pKa = 6.85FHH52 pKa = 6.91HH53 pKa = 6.59LVKK56 pKa = 10.58FPAQSSDD63 pKa = 2.86KK64 pKa = 11.28SSWDD68 pKa = 2.88WTLQPWAVDD77 pKa = 3.14IFKK80 pKa = 10.97EE81 pKa = 4.4LMLWLHH87 pKa = 6.49DD88 pKa = 3.91HH89 pKa = 6.85RR90 pKa = 11.84KK91 pKa = 8.63PWTQKK96 pKa = 9.79VMEE99 pKa = 4.06NHH101 pKa = 6.07IEE103 pKa = 3.9AMLGQKK109 pKa = 8.31TFFLRR114 pKa = 11.84GEE116 pKa = 3.97VRR118 pKa = 11.84KK119 pKa = 9.21QQDD122 pKa = 3.28RR123 pKa = 11.84GLMPSGWKK131 pKa = 7.27MTLAGNSIFQIILHH145 pKa = 5.58VLAALRR151 pKa = 11.84AGVTVSLPMTMGDD164 pKa = 3.32DD165 pKa = 3.99TVGEE169 pKa = 4.37EE170 pKa = 4.69CPPEE174 pKa = 3.71YY175 pKa = 9.83WEE177 pKa = 3.92QYY179 pKa = 9.09KK180 pKa = 10.78RR181 pKa = 11.84LGAIVKK187 pKa = 9.05EE188 pKa = 4.52VSQPSNTEE196 pKa = 3.0IDD198 pKa = 3.56FCGMIFHH205 pKa = 7.7KK206 pKa = 10.84DD207 pKa = 3.15GTFVPAYY214 pKa = 9.16KK215 pKa = 9.9EE216 pKa = 3.54KK217 pKa = 10.6HH218 pKa = 5.34AFLVRR223 pKa = 11.84RR224 pKa = 11.84IKK226 pKa = 10.87SDD228 pKa = 3.47VLKK231 pKa = 8.88EE232 pKa = 4.09TLMSYY237 pKa = 10.48QLLYY241 pKa = 11.01VYY243 pKa = 9.3DD244 pKa = 3.75QPRR247 pKa = 11.84FDD249 pKa = 6.0AIQRR253 pKa = 11.84WMRR256 pKa = 11.84SIGHH260 pKa = 6.48GSMNFEE266 pKa = 3.75RR267 pKa = 11.84EE268 pKa = 3.52QLIRR272 pKa = 11.84HH273 pKa = 5.5LQGLRR278 pKa = 11.84TKK280 pKa = 10.48RR281 pKa = 11.84RR282 pKa = 11.84FVGLSRR288 pKa = 11.84LDD290 pKa = 3.26IRR292 pKa = 5.05
MM1 pKa = 7.32WRR3 pKa = 11.84LISGVSVRR11 pKa = 11.84DD12 pKa = 3.34QVVAHH17 pKa = 5.81YY18 pKa = 10.74LFGDD22 pKa = 3.47MMEE25 pKa = 4.47VLIEE29 pKa = 4.14HH30 pKa = 7.28PGLYY34 pKa = 8.59HH35 pKa = 7.12TMIGWSPFQRR45 pKa = 11.84AGQRR49 pKa = 11.84YY50 pKa = 6.85FHH52 pKa = 6.91HH53 pKa = 6.59LVKK56 pKa = 10.58FPAQSSDD63 pKa = 2.86KK64 pKa = 11.28SSWDD68 pKa = 2.88WTLQPWAVDD77 pKa = 3.14IFKK80 pKa = 10.97EE81 pKa = 4.4LMLWLHH87 pKa = 6.49DD88 pKa = 3.91HH89 pKa = 6.85RR90 pKa = 11.84KK91 pKa = 8.63PWTQKK96 pKa = 9.79VMEE99 pKa = 4.06NHH101 pKa = 6.07IEE103 pKa = 3.9AMLGQKK109 pKa = 8.31TFFLRR114 pKa = 11.84GEE116 pKa = 3.97VRR118 pKa = 11.84KK119 pKa = 9.21QQDD122 pKa = 3.28RR123 pKa = 11.84GLMPSGWKK131 pKa = 7.27MTLAGNSIFQIILHH145 pKa = 5.58VLAALRR151 pKa = 11.84AGVTVSLPMTMGDD164 pKa = 3.32DD165 pKa = 3.99TVGEE169 pKa = 4.37EE170 pKa = 4.69CPPEE174 pKa = 3.71YY175 pKa = 9.83WEE177 pKa = 3.92QYY179 pKa = 9.09KK180 pKa = 10.78RR181 pKa = 11.84LGAIVKK187 pKa = 9.05EE188 pKa = 4.52VSQPSNTEE196 pKa = 3.0IDD198 pKa = 3.56FCGMIFHH205 pKa = 7.7KK206 pKa = 10.84DD207 pKa = 3.15GTFVPAYY214 pKa = 9.16KK215 pKa = 9.9EE216 pKa = 3.54KK217 pKa = 10.6HH218 pKa = 5.34AFLVRR223 pKa = 11.84RR224 pKa = 11.84IKK226 pKa = 10.87SDD228 pKa = 3.47VLKK231 pKa = 8.88EE232 pKa = 4.09TLMSYY237 pKa = 10.48QLLYY241 pKa = 11.01VYY243 pKa = 9.3DD244 pKa = 3.75QPRR247 pKa = 11.84FDD249 pKa = 6.0AIQRR253 pKa = 11.84WMRR256 pKa = 11.84SIGHH260 pKa = 6.48GSMNFEE266 pKa = 3.75RR267 pKa = 11.84EE268 pKa = 3.52QLIRR272 pKa = 11.84HH273 pKa = 5.5LQGLRR278 pKa = 11.84TKK280 pKa = 10.48RR281 pKa = 11.84RR282 pKa = 11.84FVGLSRR288 pKa = 11.84LDD290 pKa = 3.26IRR292 pKa = 5.05
Molecular weight: 34.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1106 |
292 |
814 |
553.0 |
62.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.6 ± 1.158 | 0.995 ± 0.167 |
4.882 ± 0.137 | 6.42 ± 0.507 |
3.436 ± 0.917 | 6.962 ± 0.061 |
2.712 ± 0.938 | 5.154 ± 0.176 |
6.51 ± 0.556 | 7.957 ± 0.88 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.521 ± 0.332 | 3.526 ± 1.163 |
4.521 ± 0.222 | 5.244 ± 0.312 |
5.515 ± 1.089 | 9.132 ± 1.785 |
5.154 ± 0.563 | 5.606 ± 0.671 |
1.899 ± 0.823 | 3.255 ± 0.093 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
