
Chalara elegans RNA Virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6Q425|Q6Q425_9VIRU Coat protein OS=Chalara elegans RNA Virus 1 OX=267285 PE=4 SV=1
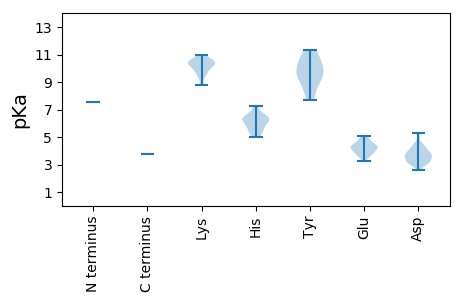
MM1 pKa = 7.58EE2 pKa = 4.73ATSLNHH8 pKa = 6.28FLSGVIAAPKK18 pKa = 9.75GGRR21 pKa = 11.84LAEE24 pKa = 4.35DD25 pKa = 3.44SAFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.13RR32 pKa = 11.84AHH34 pKa = 6.3VRR36 pKa = 11.84TTATIGGNTDD46 pKa = 2.62ARR48 pKa = 11.84NAFIRR53 pKa = 11.84YY54 pKa = 7.66EE55 pKa = 4.13VGRR58 pKa = 11.84SSGKK62 pKa = 10.18RR63 pKa = 11.84GQLLAAPPDD72 pKa = 3.54DD73 pKa = 3.58SRR75 pKa = 11.84RR76 pKa = 11.84IEE78 pKa = 3.95ASYY81 pKa = 8.29PTNAVLAEE89 pKa = 4.51DD90 pKa = 4.25FLGLAKK96 pKa = 10.32KK97 pKa = 10.38YY98 pKa = 10.88SNFSAQFQFSSLAAVAEE115 pKa = 4.22RR116 pKa = 11.84LAKK119 pKa = 10.34GLAVHH124 pKa = 6.44ATVADD129 pKa = 3.73VDD131 pKa = 4.57SVALRR136 pKa = 11.84GGAPLIVAGLGTYY149 pKa = 9.96DD150 pKa = 3.84GPINSLISSVFIPRR164 pKa = 11.84LVNNVLTGDD173 pKa = 3.96VFSVLANAIAGEE185 pKa = 4.34GASIATDD192 pKa = 3.04IAEE195 pKa = 4.66IDD197 pKa = 3.59PSTRR201 pKa = 11.84QPVIPEE207 pKa = 3.61VDD209 pKa = 2.93GDD211 pKa = 4.3GFAKK215 pKa = 10.57AATEE219 pKa = 3.73ALRR222 pKa = 11.84IVGANMAQSDD232 pKa = 3.73QGLLFSLAVTRR243 pKa = 11.84GIHH246 pKa = 6.41SVVSWLPTAMKK257 pKa = 10.28AALLGICCVWTFQCPFVGFITGSKK281 pKa = 9.69LHH283 pKa = 6.6GPPSLLSTVSRR294 pKa = 11.84SVLLCRR300 pKa = 11.84CHH302 pKa = 6.17CTDD305 pKa = 3.73PAAAVAPSDD314 pKa = 3.76PGEE317 pKa = 4.33NYY319 pKa = 10.15NGEE322 pKa = 4.24WFPTFYY328 pKa = 10.8SGTKK332 pKa = 10.37GGDD335 pKa = 3.15PTVRR339 pKa = 11.84PGGDD343 pKa = 3.21CPGNSEE349 pKa = 3.84TSGRR353 pKa = 11.84IRR355 pKa = 11.84SQLLADD361 pKa = 3.76CEE363 pKa = 4.46KK364 pKa = 10.61FFRR367 pKa = 11.84NYY369 pKa = 9.52IPALGRR375 pKa = 11.84IFGLTGSPNLAVTVAVGMSRR395 pKa = 11.84FLHH398 pKa = 6.47ADD400 pKa = 3.35PRR402 pKa = 11.84HH403 pKa = 5.52LRR405 pKa = 11.84YY406 pKa = 10.1ASVAPWYY413 pKa = 9.16WIEE416 pKa = 4.11PTSLLPSDD424 pKa = 4.32FLGSVAEE431 pKa = 4.3MNGSGSFGGKK441 pKa = 10.05DD442 pKa = 3.21STKK445 pKa = 10.67TKK447 pKa = 10.42LAWEE451 pKa = 5.06DD452 pKa = 3.22IEE454 pKa = 5.09LAGDD458 pKa = 3.54RR459 pKa = 11.84DD460 pKa = 4.4TTFSAYY466 pKa = 9.51RR467 pKa = 11.84AKK469 pKa = 10.46FLSPRR474 pKa = 11.84RR475 pKa = 11.84AWFMAHH481 pKa = 6.06WNGHH485 pKa = 5.58PDD487 pKa = 3.35NGLGCIRR494 pKa = 11.84LRR496 pKa = 11.84QADD499 pKa = 4.1PNGFIHH505 pKa = 7.26PGRR508 pKa = 11.84GTSGADD514 pKa = 3.07LRR516 pKa = 11.84DD517 pKa = 3.75RR518 pKa = 11.84LEE520 pKa = 4.98DD521 pKa = 4.0DD522 pKa = 5.15APISDD527 pKa = 4.25YY528 pKa = 11.34LWDD531 pKa = 4.52RR532 pKa = 11.84GQSPFCAPGEE542 pKa = 3.85LLNLGSTIGFLVRR555 pKa = 11.84HH556 pKa = 5.21VTFDD560 pKa = 3.76DD561 pKa = 4.27DD562 pKa = 5.27GIPTTEE568 pKa = 4.18HH569 pKa = 6.1VPTSRR574 pKa = 11.84EE575 pKa = 3.93FLDD578 pKa = 3.47TTVTIEE584 pKa = 3.67VGRR587 pKa = 11.84PLSISIGKK595 pKa = 8.79SNCPDD600 pKa = 3.0SKK602 pKa = 10.99ARR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84TRR609 pKa = 11.84ATIEE613 pKa = 3.63LGAASRR619 pKa = 11.84RR620 pKa = 11.84ARR622 pKa = 11.84AFGSAAVAEE631 pKa = 4.61MPTLSTAPRR640 pKa = 11.84ALTFAPSRR648 pKa = 11.84LMVKK652 pKa = 9.34DD653 pKa = 3.36TGGGAGSSRR662 pKa = 11.84HH663 pKa = 5.65GADD666 pKa = 3.19SGGSGADD673 pKa = 3.34DD674 pKa = 3.94AARR677 pKa = 11.84KK678 pKa = 9.7ADD680 pKa = 3.83GVPVRR685 pKa = 11.84ATAHH689 pKa = 5.02NQPVRR694 pKa = 11.84FPTLANPTGPTRR706 pKa = 11.84SSAAPAAAARR716 pKa = 11.84DD717 pKa = 3.83QSAGGPSVDD726 pKa = 3.17IVRR729 pKa = 11.84SGGDD733 pKa = 2.99AASEE737 pKa = 4.35VGSDD741 pKa = 3.76VAPAPAPGVEE751 pKa = 4.2SGVVSSSVAAPTNEE765 pKa = 3.24GRR767 pKa = 11.84NDD769 pKa = 3.16VV770 pKa = 3.77
MM1 pKa = 7.58EE2 pKa = 4.73ATSLNHH8 pKa = 6.28FLSGVIAAPKK18 pKa = 9.75GGRR21 pKa = 11.84LAEE24 pKa = 4.35DD25 pKa = 3.44SAFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.13RR32 pKa = 11.84AHH34 pKa = 6.3VRR36 pKa = 11.84TTATIGGNTDD46 pKa = 2.62ARR48 pKa = 11.84NAFIRR53 pKa = 11.84YY54 pKa = 7.66EE55 pKa = 4.13VGRR58 pKa = 11.84SSGKK62 pKa = 10.18RR63 pKa = 11.84GQLLAAPPDD72 pKa = 3.54DD73 pKa = 3.58SRR75 pKa = 11.84RR76 pKa = 11.84IEE78 pKa = 3.95ASYY81 pKa = 8.29PTNAVLAEE89 pKa = 4.51DD90 pKa = 4.25FLGLAKK96 pKa = 10.32KK97 pKa = 10.38YY98 pKa = 10.88SNFSAQFQFSSLAAVAEE115 pKa = 4.22RR116 pKa = 11.84LAKK119 pKa = 10.34GLAVHH124 pKa = 6.44ATVADD129 pKa = 3.73VDD131 pKa = 4.57SVALRR136 pKa = 11.84GGAPLIVAGLGTYY149 pKa = 9.96DD150 pKa = 3.84GPINSLISSVFIPRR164 pKa = 11.84LVNNVLTGDD173 pKa = 3.96VFSVLANAIAGEE185 pKa = 4.34GASIATDD192 pKa = 3.04IAEE195 pKa = 4.66IDD197 pKa = 3.59PSTRR201 pKa = 11.84QPVIPEE207 pKa = 3.61VDD209 pKa = 2.93GDD211 pKa = 4.3GFAKK215 pKa = 10.57AATEE219 pKa = 3.73ALRR222 pKa = 11.84IVGANMAQSDD232 pKa = 3.73QGLLFSLAVTRR243 pKa = 11.84GIHH246 pKa = 6.41SVVSWLPTAMKK257 pKa = 10.28AALLGICCVWTFQCPFVGFITGSKK281 pKa = 9.69LHH283 pKa = 6.6GPPSLLSTVSRR294 pKa = 11.84SVLLCRR300 pKa = 11.84CHH302 pKa = 6.17CTDD305 pKa = 3.73PAAAVAPSDD314 pKa = 3.76PGEE317 pKa = 4.33NYY319 pKa = 10.15NGEE322 pKa = 4.24WFPTFYY328 pKa = 10.8SGTKK332 pKa = 10.37GGDD335 pKa = 3.15PTVRR339 pKa = 11.84PGGDD343 pKa = 3.21CPGNSEE349 pKa = 3.84TSGRR353 pKa = 11.84IRR355 pKa = 11.84SQLLADD361 pKa = 3.76CEE363 pKa = 4.46KK364 pKa = 10.61FFRR367 pKa = 11.84NYY369 pKa = 9.52IPALGRR375 pKa = 11.84IFGLTGSPNLAVTVAVGMSRR395 pKa = 11.84FLHH398 pKa = 6.47ADD400 pKa = 3.35PRR402 pKa = 11.84HH403 pKa = 5.52LRR405 pKa = 11.84YY406 pKa = 10.1ASVAPWYY413 pKa = 9.16WIEE416 pKa = 4.11PTSLLPSDD424 pKa = 4.32FLGSVAEE431 pKa = 4.3MNGSGSFGGKK441 pKa = 10.05DD442 pKa = 3.21STKK445 pKa = 10.67TKK447 pKa = 10.42LAWEE451 pKa = 5.06DD452 pKa = 3.22IEE454 pKa = 5.09LAGDD458 pKa = 3.54RR459 pKa = 11.84DD460 pKa = 4.4TTFSAYY466 pKa = 9.51RR467 pKa = 11.84AKK469 pKa = 10.46FLSPRR474 pKa = 11.84RR475 pKa = 11.84AWFMAHH481 pKa = 6.06WNGHH485 pKa = 5.58PDD487 pKa = 3.35NGLGCIRR494 pKa = 11.84LRR496 pKa = 11.84QADD499 pKa = 4.1PNGFIHH505 pKa = 7.26PGRR508 pKa = 11.84GTSGADD514 pKa = 3.07LRR516 pKa = 11.84DD517 pKa = 3.75RR518 pKa = 11.84LEE520 pKa = 4.98DD521 pKa = 4.0DD522 pKa = 5.15APISDD527 pKa = 4.25YY528 pKa = 11.34LWDD531 pKa = 4.52RR532 pKa = 11.84GQSPFCAPGEE542 pKa = 3.85LLNLGSTIGFLVRR555 pKa = 11.84HH556 pKa = 5.21VTFDD560 pKa = 3.76DD561 pKa = 4.27DD562 pKa = 5.27GIPTTEE568 pKa = 4.18HH569 pKa = 6.1VPTSRR574 pKa = 11.84EE575 pKa = 3.93FLDD578 pKa = 3.47TTVTIEE584 pKa = 3.67VGRR587 pKa = 11.84PLSISIGKK595 pKa = 8.79SNCPDD600 pKa = 3.0SKK602 pKa = 10.99ARR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84TRR609 pKa = 11.84ATIEE613 pKa = 3.63LGAASRR619 pKa = 11.84RR620 pKa = 11.84ARR622 pKa = 11.84AFGSAAVAEE631 pKa = 4.61MPTLSTAPRR640 pKa = 11.84ALTFAPSRR648 pKa = 11.84LMVKK652 pKa = 9.34DD653 pKa = 3.36TGGGAGSSRR662 pKa = 11.84HH663 pKa = 5.65GADD666 pKa = 3.19SGGSGADD673 pKa = 3.34DD674 pKa = 3.94AARR677 pKa = 11.84KK678 pKa = 9.7ADD680 pKa = 3.83GVPVRR685 pKa = 11.84ATAHH689 pKa = 5.02NQPVRR694 pKa = 11.84FPTLANPTGPTRR706 pKa = 11.84SSAAPAAAARR716 pKa = 11.84DD717 pKa = 3.83QSAGGPSVDD726 pKa = 3.17IVRR729 pKa = 11.84SGGDD733 pKa = 2.99AASEE737 pKa = 4.35VGSDD741 pKa = 3.76VAPAPAPGVEE751 pKa = 4.2SGVVSSSVAAPTNEE765 pKa = 3.24GRR767 pKa = 11.84NDD769 pKa = 3.16VV770 pKa = 3.77
Molecular weight: 80.7 kDa
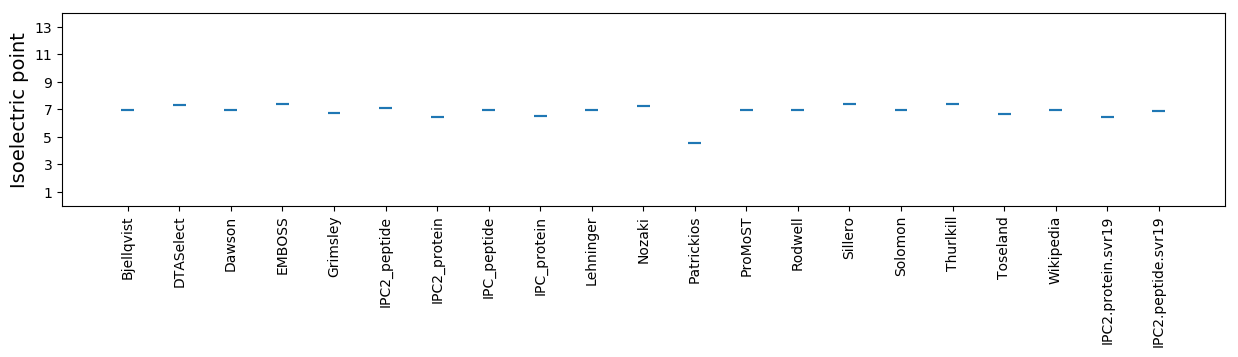
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6Q425|Q6Q425_9VIRU Coat protein OS=Chalara elegans RNA Virus 1 OX=267285 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 4.73ATSLNHH8 pKa = 6.28FLSGVIAAPKK18 pKa = 9.75GGRR21 pKa = 11.84LAEE24 pKa = 4.35DD25 pKa = 3.44SAFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.13RR32 pKa = 11.84AHH34 pKa = 6.3VRR36 pKa = 11.84TTATIGGNTDD46 pKa = 2.62ARR48 pKa = 11.84NAFIRR53 pKa = 11.84YY54 pKa = 7.66EE55 pKa = 4.13VGRR58 pKa = 11.84SSGKK62 pKa = 10.18RR63 pKa = 11.84GQLLAAPPDD72 pKa = 3.54DD73 pKa = 3.58SRR75 pKa = 11.84RR76 pKa = 11.84IEE78 pKa = 3.95ASYY81 pKa = 8.29PTNAVLAEE89 pKa = 4.51DD90 pKa = 4.25FLGLAKK96 pKa = 10.32KK97 pKa = 10.38YY98 pKa = 10.88SNFSAQFQFSSLAAVAEE115 pKa = 4.22RR116 pKa = 11.84LAKK119 pKa = 10.34GLAVHH124 pKa = 6.44ATVADD129 pKa = 3.73VDD131 pKa = 4.57SVALRR136 pKa = 11.84GGAPLIVAGLGTYY149 pKa = 9.96DD150 pKa = 3.84GPINSLISSVFIPRR164 pKa = 11.84LVNNVLTGDD173 pKa = 3.96VFSVLANAIAGEE185 pKa = 4.34GASIATDD192 pKa = 3.04IAEE195 pKa = 4.66IDD197 pKa = 3.59PSTRR201 pKa = 11.84QPVIPEE207 pKa = 3.61VDD209 pKa = 2.93GDD211 pKa = 4.3GFAKK215 pKa = 10.57AATEE219 pKa = 3.73ALRR222 pKa = 11.84IVGANMAQSDD232 pKa = 3.73QGLLFSLAVTRR243 pKa = 11.84GIHH246 pKa = 6.41SVVSWLPTAMKK257 pKa = 10.28AALLGICCVWTFQCPFVGFITGSKK281 pKa = 9.69LHH283 pKa = 6.6GPPSLLSTVSRR294 pKa = 11.84SVLLCRR300 pKa = 11.84CHH302 pKa = 6.17CTDD305 pKa = 3.73PAAAVAPSDD314 pKa = 3.76PGEE317 pKa = 4.33NYY319 pKa = 10.15NGEE322 pKa = 4.24WFPTFYY328 pKa = 10.8SGTKK332 pKa = 10.37GGDD335 pKa = 3.15PTVRR339 pKa = 11.84PGGDD343 pKa = 3.21CPGNSEE349 pKa = 3.84TSGRR353 pKa = 11.84IRR355 pKa = 11.84SQLLADD361 pKa = 3.76CEE363 pKa = 4.46KK364 pKa = 10.61FFRR367 pKa = 11.84NYY369 pKa = 9.52IPALGRR375 pKa = 11.84IFGLTGSPNLAVTVAVGMSRR395 pKa = 11.84FLHH398 pKa = 6.47ADD400 pKa = 3.35PRR402 pKa = 11.84HH403 pKa = 5.52LRR405 pKa = 11.84YY406 pKa = 10.1ASVAPWYY413 pKa = 9.16WIEE416 pKa = 4.11PTSLLPSDD424 pKa = 4.32FLGSVAEE431 pKa = 4.3MNGSGSFGGKK441 pKa = 10.05DD442 pKa = 3.21STKK445 pKa = 10.67TKK447 pKa = 10.42LAWEE451 pKa = 5.06DD452 pKa = 3.22IEE454 pKa = 5.09LAGDD458 pKa = 3.54RR459 pKa = 11.84DD460 pKa = 4.4TTFSAYY466 pKa = 9.51RR467 pKa = 11.84AKK469 pKa = 10.46FLSPRR474 pKa = 11.84RR475 pKa = 11.84AWFMAHH481 pKa = 6.06WNGHH485 pKa = 5.58PDD487 pKa = 3.35NGLGCIRR494 pKa = 11.84LRR496 pKa = 11.84QADD499 pKa = 4.1PNGFIHH505 pKa = 7.26PGRR508 pKa = 11.84GTSGADD514 pKa = 3.07LRR516 pKa = 11.84DD517 pKa = 3.75RR518 pKa = 11.84LEE520 pKa = 4.98DD521 pKa = 4.0DD522 pKa = 5.15APISDD527 pKa = 4.25YY528 pKa = 11.34LWDD531 pKa = 4.52RR532 pKa = 11.84GQSPFCAPGEE542 pKa = 3.85LLNLGSTIGFLVRR555 pKa = 11.84HH556 pKa = 5.21VTFDD560 pKa = 3.76DD561 pKa = 4.27DD562 pKa = 5.27GIPTTEE568 pKa = 4.18HH569 pKa = 6.1VPTSRR574 pKa = 11.84EE575 pKa = 3.93FLDD578 pKa = 3.47TTVTIEE584 pKa = 3.67VGRR587 pKa = 11.84PLSISIGKK595 pKa = 8.79SNCPDD600 pKa = 3.0SKK602 pKa = 10.99ARR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84TRR609 pKa = 11.84ATIEE613 pKa = 3.63LGAASRR619 pKa = 11.84RR620 pKa = 11.84ARR622 pKa = 11.84AFGSAAVAEE631 pKa = 4.61MPTLSTAPRR640 pKa = 11.84ALTFAPSRR648 pKa = 11.84LMVKK652 pKa = 9.34DD653 pKa = 3.36TGGGAGSSRR662 pKa = 11.84HH663 pKa = 5.65GADD666 pKa = 3.19SGGSGADD673 pKa = 3.34DD674 pKa = 3.94AARR677 pKa = 11.84KK678 pKa = 9.7ADD680 pKa = 3.83GVPVRR685 pKa = 11.84ATAHH689 pKa = 5.02NQPVRR694 pKa = 11.84FPTLANPTGPTRR706 pKa = 11.84SSAAPAAAARR716 pKa = 11.84DD717 pKa = 3.83QSAGGPSVDD726 pKa = 3.17IVRR729 pKa = 11.84SGGDD733 pKa = 2.99AASEE737 pKa = 4.35VGSDD741 pKa = 3.76VAPAPAPGVEE751 pKa = 4.2SGVVSSSVAAPTNEE765 pKa = 3.24GRR767 pKa = 11.84NDD769 pKa = 3.16VV770 pKa = 3.77
MM1 pKa = 7.58EE2 pKa = 4.73ATSLNHH8 pKa = 6.28FLSGVIAAPKK18 pKa = 9.75GGRR21 pKa = 11.84LAEE24 pKa = 4.35DD25 pKa = 3.44SAFRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 9.13RR32 pKa = 11.84AHH34 pKa = 6.3VRR36 pKa = 11.84TTATIGGNTDD46 pKa = 2.62ARR48 pKa = 11.84NAFIRR53 pKa = 11.84YY54 pKa = 7.66EE55 pKa = 4.13VGRR58 pKa = 11.84SSGKK62 pKa = 10.18RR63 pKa = 11.84GQLLAAPPDD72 pKa = 3.54DD73 pKa = 3.58SRR75 pKa = 11.84RR76 pKa = 11.84IEE78 pKa = 3.95ASYY81 pKa = 8.29PTNAVLAEE89 pKa = 4.51DD90 pKa = 4.25FLGLAKK96 pKa = 10.32KK97 pKa = 10.38YY98 pKa = 10.88SNFSAQFQFSSLAAVAEE115 pKa = 4.22RR116 pKa = 11.84LAKK119 pKa = 10.34GLAVHH124 pKa = 6.44ATVADD129 pKa = 3.73VDD131 pKa = 4.57SVALRR136 pKa = 11.84GGAPLIVAGLGTYY149 pKa = 9.96DD150 pKa = 3.84GPINSLISSVFIPRR164 pKa = 11.84LVNNVLTGDD173 pKa = 3.96VFSVLANAIAGEE185 pKa = 4.34GASIATDD192 pKa = 3.04IAEE195 pKa = 4.66IDD197 pKa = 3.59PSTRR201 pKa = 11.84QPVIPEE207 pKa = 3.61VDD209 pKa = 2.93GDD211 pKa = 4.3GFAKK215 pKa = 10.57AATEE219 pKa = 3.73ALRR222 pKa = 11.84IVGANMAQSDD232 pKa = 3.73QGLLFSLAVTRR243 pKa = 11.84GIHH246 pKa = 6.41SVVSWLPTAMKK257 pKa = 10.28AALLGICCVWTFQCPFVGFITGSKK281 pKa = 9.69LHH283 pKa = 6.6GPPSLLSTVSRR294 pKa = 11.84SVLLCRR300 pKa = 11.84CHH302 pKa = 6.17CTDD305 pKa = 3.73PAAAVAPSDD314 pKa = 3.76PGEE317 pKa = 4.33NYY319 pKa = 10.15NGEE322 pKa = 4.24WFPTFYY328 pKa = 10.8SGTKK332 pKa = 10.37GGDD335 pKa = 3.15PTVRR339 pKa = 11.84PGGDD343 pKa = 3.21CPGNSEE349 pKa = 3.84TSGRR353 pKa = 11.84IRR355 pKa = 11.84SQLLADD361 pKa = 3.76CEE363 pKa = 4.46KK364 pKa = 10.61FFRR367 pKa = 11.84NYY369 pKa = 9.52IPALGRR375 pKa = 11.84IFGLTGSPNLAVTVAVGMSRR395 pKa = 11.84FLHH398 pKa = 6.47ADD400 pKa = 3.35PRR402 pKa = 11.84HH403 pKa = 5.52LRR405 pKa = 11.84YY406 pKa = 10.1ASVAPWYY413 pKa = 9.16WIEE416 pKa = 4.11PTSLLPSDD424 pKa = 4.32FLGSVAEE431 pKa = 4.3MNGSGSFGGKK441 pKa = 10.05DD442 pKa = 3.21STKK445 pKa = 10.67TKK447 pKa = 10.42LAWEE451 pKa = 5.06DD452 pKa = 3.22IEE454 pKa = 5.09LAGDD458 pKa = 3.54RR459 pKa = 11.84DD460 pKa = 4.4TTFSAYY466 pKa = 9.51RR467 pKa = 11.84AKK469 pKa = 10.46FLSPRR474 pKa = 11.84RR475 pKa = 11.84AWFMAHH481 pKa = 6.06WNGHH485 pKa = 5.58PDD487 pKa = 3.35NGLGCIRR494 pKa = 11.84LRR496 pKa = 11.84QADD499 pKa = 4.1PNGFIHH505 pKa = 7.26PGRR508 pKa = 11.84GTSGADD514 pKa = 3.07LRR516 pKa = 11.84DD517 pKa = 3.75RR518 pKa = 11.84LEE520 pKa = 4.98DD521 pKa = 4.0DD522 pKa = 5.15APISDD527 pKa = 4.25YY528 pKa = 11.34LWDD531 pKa = 4.52RR532 pKa = 11.84GQSPFCAPGEE542 pKa = 3.85LLNLGSTIGFLVRR555 pKa = 11.84HH556 pKa = 5.21VTFDD560 pKa = 3.76DD561 pKa = 4.27DD562 pKa = 5.27GIPTTEE568 pKa = 4.18HH569 pKa = 6.1VPTSRR574 pKa = 11.84EE575 pKa = 3.93FLDD578 pKa = 3.47TTVTIEE584 pKa = 3.67VGRR587 pKa = 11.84PLSISIGKK595 pKa = 8.79SNCPDD600 pKa = 3.0SKK602 pKa = 10.99ARR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84TRR609 pKa = 11.84ATIEE613 pKa = 3.63LGAASRR619 pKa = 11.84RR620 pKa = 11.84ARR622 pKa = 11.84AFGSAAVAEE631 pKa = 4.61MPTLSTAPRR640 pKa = 11.84ALTFAPSRR648 pKa = 11.84LMVKK652 pKa = 9.34DD653 pKa = 3.36TGGGAGSSRR662 pKa = 11.84HH663 pKa = 5.65GADD666 pKa = 3.19SGGSGADD673 pKa = 3.34DD674 pKa = 3.94AARR677 pKa = 11.84KK678 pKa = 9.7ADD680 pKa = 3.83GVPVRR685 pKa = 11.84ATAHH689 pKa = 5.02NQPVRR694 pKa = 11.84FPTLANPTGPTRR706 pKa = 11.84SSAAPAAAARR716 pKa = 11.84DD717 pKa = 3.83QSAGGPSVDD726 pKa = 3.17IVRR729 pKa = 11.84SGGDD733 pKa = 2.99AASEE737 pKa = 4.35VGSDD741 pKa = 3.76VAPAPAPGVEE751 pKa = 4.2SGVVSSSVAAPTNEE765 pKa = 3.24GRR767 pKa = 11.84NDD769 pKa = 3.16VV770 pKa = 3.77
Molecular weight: 80.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
770 |
770 |
770 |
770.0 |
80.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.987 ± 0.0 | 1.429 ± 0.0 |
6.234 ± 0.0 | 3.636 ± 0.0 |
4.286 ± 0.0 | 10.39 ± 0.0 |
1.948 ± 0.0 | 4.156 ± 0.0 |
2.338 ± 0.0 | 7.662 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.039 ± 0.0 | 3.247 ± 0.0 |
6.753 ± 0.0 | 1.558 ± 0.0 |
7.403 ± 0.0 | 9.221 ± 0.0 |
6.364 ± 0.0 | 6.623 ± 0.0 |
1.169 ± 0.0 | 1.558 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
