
Rosellinia necatrix partitivirus 8
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
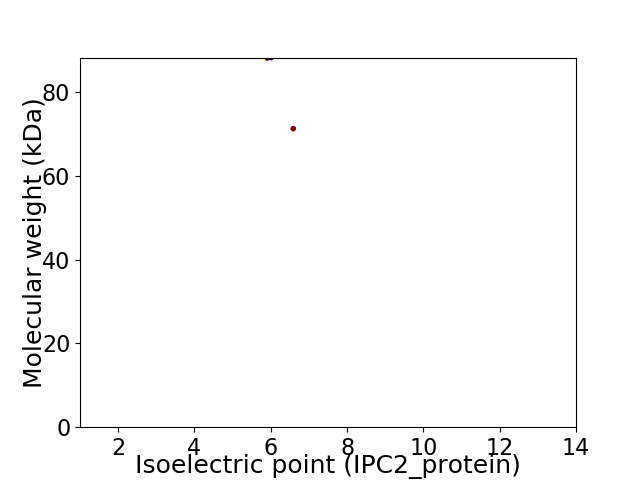
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
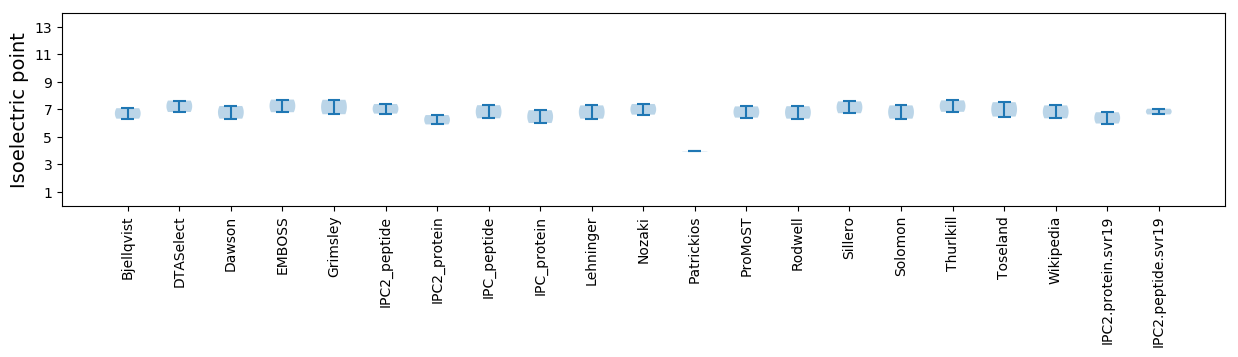
Protein with the lowest isoelectric point:
>tr|A0A2Z5WQE2|A0A2Z5WQE2_9VIRU Coat protein OS=Rosellinia necatrix partitivirus 8 OX=2025333 PE=4 SV=1
MM1 pKa = 7.27LTLFTIVTLNILLRR15 pKa = 11.84FQQRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84NEE24 pKa = 3.77TTSTLIHH31 pKa = 7.26PIRR34 pKa = 11.84NILFEE39 pKa = 4.14WEE41 pKa = 4.17FRR43 pKa = 11.84RR44 pKa = 11.84SIPLDD49 pKa = 3.52ASPYY53 pKa = 9.98PVFDD57 pKa = 3.65YY58 pKa = 11.13SKK60 pKa = 11.3SLLAYY65 pKa = 9.29IDD67 pKa = 5.96LITCHH72 pKa = 6.95KK73 pKa = 10.37ISIHH77 pKa = 5.76LKK79 pKa = 8.19EE80 pKa = 5.45DD81 pKa = 3.06IYY83 pKa = 11.09TLEE86 pKa = 3.9EE87 pKa = 4.45RR88 pKa = 11.84YY89 pKa = 10.06SSRR92 pKa = 11.84DD93 pKa = 3.54EE94 pKa = 4.32NFHH97 pKa = 7.4LYY99 pKa = 10.37EE100 pKa = 4.75YY101 pKa = 9.62ITDD104 pKa = 3.94ADD106 pKa = 4.57LPPDD110 pKa = 3.76RR111 pKa = 11.84QPANGISHH119 pKa = 7.29AEE121 pKa = 3.87RR122 pKa = 11.84RR123 pKa = 11.84YY124 pKa = 10.16HH125 pKa = 6.86SIPSGSLNVNDD136 pKa = 4.8KK137 pKa = 11.12IEE139 pKa = 4.48VISNDD144 pKa = 3.17PDD146 pKa = 4.33FIEE149 pKa = 4.45TNEE152 pKa = 3.79FRR154 pKa = 11.84SNILFAEE161 pKa = 4.28SLDD164 pKa = 3.93LTGSPPHH171 pKa = 5.98PQIEE175 pKa = 4.62KK176 pKa = 10.62LIQDD180 pKa = 4.4WFPQYY185 pKa = 11.08VPYY188 pKa = 10.52LHH190 pKa = 7.18EE191 pKa = 4.18YY192 pKa = 9.58CRR194 pKa = 11.84PPSFGPQAFLDD205 pKa = 4.29FNRR208 pKa = 11.84DD209 pKa = 3.35TPDD212 pKa = 3.78PQPPTPEE219 pKa = 3.39RR220 pKa = 11.84HH221 pKa = 4.71EE222 pKa = 4.64AIMRR226 pKa = 11.84IVRR229 pKa = 11.84LKK231 pKa = 10.73MNIKK235 pKa = 9.99PYY237 pKa = 10.32RR238 pKa = 11.84PLHH241 pKa = 5.77FADD244 pKa = 4.26ALAAEE249 pKa = 4.81TPLNTSASYY258 pKa = 10.43YY259 pKa = 10.74SKK261 pKa = 10.79FNPEE265 pKa = 3.44TRR267 pKa = 11.84VLARR271 pKa = 11.84YY272 pKa = 7.4STPSRR277 pKa = 11.84YY278 pKa = 10.09SDD280 pKa = 3.13MPTSKK285 pKa = 10.81GYY287 pKa = 10.45FINVMLNEE295 pKa = 3.9FRR297 pKa = 11.84LEE299 pKa = 3.91FHH301 pKa = 6.88HH302 pKa = 6.85IKK304 pKa = 10.15YY305 pKa = 10.53DD306 pKa = 3.6GMPFPTDD313 pKa = 3.06RR314 pKa = 11.84HH315 pKa = 5.19DD316 pKa = 4.02HH317 pKa = 5.35EE318 pKa = 5.12TNLTILDD325 pKa = 3.49TWLAKK330 pKa = 10.5HH331 pKa = 6.27PAQLFIRR338 pKa = 11.84TQISKK343 pKa = 10.3RR344 pKa = 11.84DD345 pKa = 3.39PSDD348 pKa = 3.26PKK350 pKa = 10.5KK351 pKa = 10.23IRR353 pKa = 11.84PVYY356 pKa = 10.59SVDD359 pKa = 4.16DD360 pKa = 3.94RR361 pKa = 11.84FLHH364 pKa = 6.15IEE366 pKa = 4.2KK367 pKa = 9.54TVSTPLLAQMRR378 pKa = 11.84NPQCCVAHH386 pKa = 5.99GLEE389 pKa = 4.28TFRR392 pKa = 11.84GSMSLIDD399 pKa = 4.4KK400 pKa = 9.63IAHH403 pKa = 6.37FFLSFISLDD412 pKa = 2.89WSQFDD417 pKa = 3.23QRR419 pKa = 11.84LPYY422 pKa = 10.14YY423 pKa = 10.23VIIAFFLDD431 pKa = 4.4FLASLIIVSHH441 pKa = 6.7GYY443 pKa = 8.53MPTRR447 pKa = 11.84SYY449 pKa = 11.39PDD451 pKa = 3.37TKK453 pKa = 11.21SDD455 pKa = 2.69IHH457 pKa = 7.82SFARR461 pKa = 11.84RR462 pKa = 11.84QYY464 pKa = 9.77NVLVFLTTWYY474 pKa = 11.15LSMTFLSFDD483 pKa = 3.03GFAFIRR489 pKa = 11.84IHH491 pKa = 6.33GGVPSGLLNTQSLDD505 pKa = 3.37SFGNMYY511 pKa = 10.44IITDD515 pKa = 3.8CLLEE519 pKa = 4.23FGFTEE524 pKa = 4.41QEE526 pKa = 4.07CLEE529 pKa = 4.16MLFCVLGDD537 pKa = 3.91DD538 pKa = 4.4NLIFLQHH545 pKa = 5.62NLEE548 pKa = 4.78RR549 pKa = 11.84ITRR552 pKa = 11.84FMVFLEE558 pKa = 4.63RR559 pKa = 11.84YY560 pKa = 8.88SKK562 pKa = 10.55DD563 pKa = 2.83RR564 pKa = 11.84HH565 pKa = 5.9GMVLSILKK573 pKa = 10.83SMFSNLRR580 pKa = 11.84TKK582 pKa = 9.21ITFLSYY588 pKa = 11.31EE589 pKa = 4.09NFFGHH594 pKa = 7.23PSRR597 pKa = 11.84PIGKK601 pKa = 9.18LVAQLAFPEE610 pKa = 4.64RR611 pKa = 11.84PVPPARR617 pKa = 11.84EE618 pKa = 4.11WIHH621 pKa = 6.13AARR624 pKa = 11.84ALGLAYY630 pKa = 9.97ASCGQDD636 pKa = 3.01PTFHH640 pKa = 7.23LLCKK644 pKa = 9.53MVYY647 pKa = 9.14EE648 pKa = 4.55RR649 pKa = 11.84FRR651 pKa = 11.84PSVPVPTLHH660 pKa = 6.77INKK663 pKa = 9.03IFKK666 pKa = 8.89KK667 pKa = 9.44WKK669 pKa = 8.48YY670 pKa = 9.07QLPDD674 pKa = 3.53FDD676 pKa = 5.05IEE678 pKa = 4.12EE679 pKa = 4.44LEE681 pKa = 4.24YY682 pKa = 10.74TFPDD686 pKa = 4.69FPTCTEE692 pKa = 3.67IFALVSDD699 pKa = 3.82YY700 pKa = 11.39HH701 pKa = 8.12GVFSEE706 pKa = 4.09TDD708 pKa = 2.5KK709 pKa = 11.33WNFNVFTVPPSDD721 pKa = 3.88NLPDD725 pKa = 3.67YY726 pKa = 9.74VTLKK730 pKa = 10.96NYY732 pKa = 9.15IQQSPDD738 pKa = 2.74VSHH741 pKa = 6.47TVNEE745 pKa = 4.48FWHH748 pKa = 6.46GKK750 pKa = 9.82RR751 pKa = 11.84PFF753 pKa = 3.6
MM1 pKa = 7.27LTLFTIVTLNILLRR15 pKa = 11.84FQQRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84NEE24 pKa = 3.77TTSTLIHH31 pKa = 7.26PIRR34 pKa = 11.84NILFEE39 pKa = 4.14WEE41 pKa = 4.17FRR43 pKa = 11.84RR44 pKa = 11.84SIPLDD49 pKa = 3.52ASPYY53 pKa = 9.98PVFDD57 pKa = 3.65YY58 pKa = 11.13SKK60 pKa = 11.3SLLAYY65 pKa = 9.29IDD67 pKa = 5.96LITCHH72 pKa = 6.95KK73 pKa = 10.37ISIHH77 pKa = 5.76LKK79 pKa = 8.19EE80 pKa = 5.45DD81 pKa = 3.06IYY83 pKa = 11.09TLEE86 pKa = 3.9EE87 pKa = 4.45RR88 pKa = 11.84YY89 pKa = 10.06SSRR92 pKa = 11.84DD93 pKa = 3.54EE94 pKa = 4.32NFHH97 pKa = 7.4LYY99 pKa = 10.37EE100 pKa = 4.75YY101 pKa = 9.62ITDD104 pKa = 3.94ADD106 pKa = 4.57LPPDD110 pKa = 3.76RR111 pKa = 11.84QPANGISHH119 pKa = 7.29AEE121 pKa = 3.87RR122 pKa = 11.84RR123 pKa = 11.84YY124 pKa = 10.16HH125 pKa = 6.86SIPSGSLNVNDD136 pKa = 4.8KK137 pKa = 11.12IEE139 pKa = 4.48VISNDD144 pKa = 3.17PDD146 pKa = 4.33FIEE149 pKa = 4.45TNEE152 pKa = 3.79FRR154 pKa = 11.84SNILFAEE161 pKa = 4.28SLDD164 pKa = 3.93LTGSPPHH171 pKa = 5.98PQIEE175 pKa = 4.62KK176 pKa = 10.62LIQDD180 pKa = 4.4WFPQYY185 pKa = 11.08VPYY188 pKa = 10.52LHH190 pKa = 7.18EE191 pKa = 4.18YY192 pKa = 9.58CRR194 pKa = 11.84PPSFGPQAFLDD205 pKa = 4.29FNRR208 pKa = 11.84DD209 pKa = 3.35TPDD212 pKa = 3.78PQPPTPEE219 pKa = 3.39RR220 pKa = 11.84HH221 pKa = 4.71EE222 pKa = 4.64AIMRR226 pKa = 11.84IVRR229 pKa = 11.84LKK231 pKa = 10.73MNIKK235 pKa = 9.99PYY237 pKa = 10.32RR238 pKa = 11.84PLHH241 pKa = 5.77FADD244 pKa = 4.26ALAAEE249 pKa = 4.81TPLNTSASYY258 pKa = 10.43YY259 pKa = 10.74SKK261 pKa = 10.79FNPEE265 pKa = 3.44TRR267 pKa = 11.84VLARR271 pKa = 11.84YY272 pKa = 7.4STPSRR277 pKa = 11.84YY278 pKa = 10.09SDD280 pKa = 3.13MPTSKK285 pKa = 10.81GYY287 pKa = 10.45FINVMLNEE295 pKa = 3.9FRR297 pKa = 11.84LEE299 pKa = 3.91FHH301 pKa = 6.88HH302 pKa = 6.85IKK304 pKa = 10.15YY305 pKa = 10.53DD306 pKa = 3.6GMPFPTDD313 pKa = 3.06RR314 pKa = 11.84HH315 pKa = 5.19DD316 pKa = 4.02HH317 pKa = 5.35EE318 pKa = 5.12TNLTILDD325 pKa = 3.49TWLAKK330 pKa = 10.5HH331 pKa = 6.27PAQLFIRR338 pKa = 11.84TQISKK343 pKa = 10.3RR344 pKa = 11.84DD345 pKa = 3.39PSDD348 pKa = 3.26PKK350 pKa = 10.5KK351 pKa = 10.23IRR353 pKa = 11.84PVYY356 pKa = 10.59SVDD359 pKa = 4.16DD360 pKa = 3.94RR361 pKa = 11.84FLHH364 pKa = 6.15IEE366 pKa = 4.2KK367 pKa = 9.54TVSTPLLAQMRR378 pKa = 11.84NPQCCVAHH386 pKa = 5.99GLEE389 pKa = 4.28TFRR392 pKa = 11.84GSMSLIDD399 pKa = 4.4KK400 pKa = 9.63IAHH403 pKa = 6.37FFLSFISLDD412 pKa = 2.89WSQFDD417 pKa = 3.23QRR419 pKa = 11.84LPYY422 pKa = 10.14YY423 pKa = 10.23VIIAFFLDD431 pKa = 4.4FLASLIIVSHH441 pKa = 6.7GYY443 pKa = 8.53MPTRR447 pKa = 11.84SYY449 pKa = 11.39PDD451 pKa = 3.37TKK453 pKa = 11.21SDD455 pKa = 2.69IHH457 pKa = 7.82SFARR461 pKa = 11.84RR462 pKa = 11.84QYY464 pKa = 9.77NVLVFLTTWYY474 pKa = 11.15LSMTFLSFDD483 pKa = 3.03GFAFIRR489 pKa = 11.84IHH491 pKa = 6.33GGVPSGLLNTQSLDD505 pKa = 3.37SFGNMYY511 pKa = 10.44IITDD515 pKa = 3.8CLLEE519 pKa = 4.23FGFTEE524 pKa = 4.41QEE526 pKa = 4.07CLEE529 pKa = 4.16MLFCVLGDD537 pKa = 3.91DD538 pKa = 4.4NLIFLQHH545 pKa = 5.62NLEE548 pKa = 4.78RR549 pKa = 11.84ITRR552 pKa = 11.84FMVFLEE558 pKa = 4.63RR559 pKa = 11.84YY560 pKa = 8.88SKK562 pKa = 10.55DD563 pKa = 2.83RR564 pKa = 11.84HH565 pKa = 5.9GMVLSILKK573 pKa = 10.83SMFSNLRR580 pKa = 11.84TKK582 pKa = 9.21ITFLSYY588 pKa = 11.31EE589 pKa = 4.09NFFGHH594 pKa = 7.23PSRR597 pKa = 11.84PIGKK601 pKa = 9.18LVAQLAFPEE610 pKa = 4.64RR611 pKa = 11.84PVPPARR617 pKa = 11.84EE618 pKa = 4.11WIHH621 pKa = 6.13AARR624 pKa = 11.84ALGLAYY630 pKa = 9.97ASCGQDD636 pKa = 3.01PTFHH640 pKa = 7.23LLCKK644 pKa = 9.53MVYY647 pKa = 9.14EE648 pKa = 4.55RR649 pKa = 11.84FRR651 pKa = 11.84PSVPVPTLHH660 pKa = 6.77INKK663 pKa = 9.03IFKK666 pKa = 8.89KK667 pKa = 9.44WKK669 pKa = 8.48YY670 pKa = 9.07QLPDD674 pKa = 3.53FDD676 pKa = 5.05IEE678 pKa = 4.12EE679 pKa = 4.44LEE681 pKa = 4.24YY682 pKa = 10.74TFPDD686 pKa = 4.69FPTCTEE692 pKa = 3.67IFALVSDD699 pKa = 3.82YY700 pKa = 11.39HH701 pKa = 8.12GVFSEE706 pKa = 4.09TDD708 pKa = 2.5KK709 pKa = 11.33WNFNVFTVPPSDD721 pKa = 3.88NLPDD725 pKa = 3.67YY726 pKa = 9.74VTLKK730 pKa = 10.96NYY732 pKa = 9.15IQQSPDD738 pKa = 2.74VSHH741 pKa = 6.47TVNEE745 pKa = 4.48FWHH748 pKa = 6.46GKK750 pKa = 9.82RR751 pKa = 11.84PFF753 pKa = 3.6
Molecular weight: 88.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5WQE2|A0A2Z5WQE2_9VIRU Coat protein OS=Rosellinia necatrix partitivirus 8 OX=2025333 PE=4 SV=1
MM1 pKa = 7.38SSSRR5 pKa = 11.84LRR7 pKa = 11.84STIPAPAAPTVTVQDD22 pKa = 4.1GQPAAPASVAPQVLQYY38 pKa = 11.03LDD40 pKa = 3.65PSRR43 pKa = 11.84NTDD46 pKa = 3.2SNTGILFNLEE56 pKa = 3.91WVPDD60 pKa = 4.23FRR62 pKa = 11.84FMLLSWLYY70 pKa = 9.1SLARR74 pKa = 11.84IVPNSAYY81 pKa = 9.36TSSPYY86 pKa = 10.78ASPASSIGYY95 pKa = 7.32TIVMLVAFLYY105 pKa = 8.91HH106 pKa = 6.63TDD108 pKa = 3.58ASHH111 pKa = 6.56LQHH114 pKa = 6.93PSAAASAIMNDD125 pKa = 3.14ALFSRR130 pKa = 11.84FFDD133 pKa = 3.47MLLDD137 pKa = 3.97FPVPDD142 pKa = 4.08FASNEE147 pKa = 4.15FASLMAFLPDD157 pKa = 5.52DD158 pKa = 3.87IPNLVILTSLASADD172 pKa = 3.86YY173 pKa = 8.57YY174 pKa = 11.3HH175 pKa = 7.78DD176 pKa = 4.09FGRR179 pKa = 11.84HH180 pKa = 4.02FSANIFFLAHH190 pKa = 6.42NLLAGLPANTPTSTLRR206 pKa = 11.84ATFYY210 pKa = 8.6ATPVNSVNIGNNNNVNITPGQLFGRR235 pKa = 11.84INGANTVSNWLNQRR249 pKa = 11.84IDD251 pKa = 3.32AMINSMAIRR260 pKa = 11.84AVNQNNMVAQIQFPTVALANTANYY284 pKa = 10.33NPYY287 pKa = 10.67LFLASLDD294 pKa = 3.43QHH296 pKa = 6.85NIGSITSAMRR306 pKa = 11.84NMSQWISATFPASKK320 pKa = 9.33TLRR323 pKa = 11.84HH324 pKa = 5.27YY325 pKa = 8.9MQAGSHH331 pKa = 5.01EE332 pKa = 4.17TSNYY336 pKa = 9.7LFAEE340 pKa = 4.29IALPTWNTTQITITVPVSQTNSDD363 pKa = 3.47KK364 pKa = 10.79FLPTAAPDD372 pKa = 3.5QNATEE377 pKa = 4.41LAHH380 pKa = 7.46DD381 pKa = 4.24SGFLTVPTAPADD393 pKa = 3.54TDD395 pKa = 3.96VNGTSVWNIAAEE407 pKa = 4.44RR408 pKa = 11.84AASTPPANPMYY419 pKa = 10.92ASLRR423 pKa = 11.84SSATRR428 pKa = 11.84PDD430 pKa = 4.22PYY432 pKa = 10.93VNPIPHH438 pKa = 5.48VTFDD442 pKa = 3.81VNLHH446 pKa = 5.19VLPRR450 pKa = 11.84MYY452 pKa = 10.18IYY454 pKa = 10.88APYY457 pKa = 8.5VTNTGALGSVITSGKK472 pKa = 10.0LIEE475 pKa = 5.05SGDD478 pKa = 3.4ISGIMQIVPSTTTPLLYY495 pKa = 10.6EE496 pKa = 4.04NSQFHH501 pKa = 7.33DD502 pKa = 3.86GAIRR506 pKa = 11.84LSRR509 pKa = 11.84TRR511 pKa = 11.84SALFHH516 pKa = 6.84AGIQIHH522 pKa = 4.94MQEE525 pKa = 3.99RR526 pKa = 11.84PARR529 pKa = 11.84RR530 pKa = 11.84TLDD533 pKa = 3.21APIAFFRR540 pKa = 11.84GLTSGLRR547 pKa = 11.84YY548 pKa = 8.53PVPSTGPVYY557 pKa = 10.73APSAFPNSSQARR569 pKa = 11.84LLPGARR575 pKa = 11.84WTSHH579 pKa = 4.42ATNFFHH585 pKa = 6.7TLNVFGRR592 pKa = 11.84DD593 pKa = 3.54LSHH596 pKa = 7.8DD597 pKa = 4.08LQLHH601 pKa = 6.37ADD603 pKa = 3.4SSFNIWSGLRR613 pKa = 11.84FRR615 pKa = 11.84YY616 pKa = 6.75MTNTGPVVYY625 pKa = 9.4VLPSARR631 pKa = 11.84HH632 pKa = 4.68IFGARR637 pKa = 11.84ARR639 pKa = 11.84SYY641 pKa = 9.58GTEE644 pKa = 3.97HH645 pKa = 7.07PALRR649 pKa = 11.84LPLL652 pKa = 4.15
MM1 pKa = 7.38SSSRR5 pKa = 11.84LRR7 pKa = 11.84STIPAPAAPTVTVQDD22 pKa = 4.1GQPAAPASVAPQVLQYY38 pKa = 11.03LDD40 pKa = 3.65PSRR43 pKa = 11.84NTDD46 pKa = 3.2SNTGILFNLEE56 pKa = 3.91WVPDD60 pKa = 4.23FRR62 pKa = 11.84FMLLSWLYY70 pKa = 9.1SLARR74 pKa = 11.84IVPNSAYY81 pKa = 9.36TSSPYY86 pKa = 10.78ASPASSIGYY95 pKa = 7.32TIVMLVAFLYY105 pKa = 8.91HH106 pKa = 6.63TDD108 pKa = 3.58ASHH111 pKa = 6.56LQHH114 pKa = 6.93PSAAASAIMNDD125 pKa = 3.14ALFSRR130 pKa = 11.84FFDD133 pKa = 3.47MLLDD137 pKa = 3.97FPVPDD142 pKa = 4.08FASNEE147 pKa = 4.15FASLMAFLPDD157 pKa = 5.52DD158 pKa = 3.87IPNLVILTSLASADD172 pKa = 3.86YY173 pKa = 8.57YY174 pKa = 11.3HH175 pKa = 7.78DD176 pKa = 4.09FGRR179 pKa = 11.84HH180 pKa = 4.02FSANIFFLAHH190 pKa = 6.42NLLAGLPANTPTSTLRR206 pKa = 11.84ATFYY210 pKa = 8.6ATPVNSVNIGNNNNVNITPGQLFGRR235 pKa = 11.84INGANTVSNWLNQRR249 pKa = 11.84IDD251 pKa = 3.32AMINSMAIRR260 pKa = 11.84AVNQNNMVAQIQFPTVALANTANYY284 pKa = 10.33NPYY287 pKa = 10.67LFLASLDD294 pKa = 3.43QHH296 pKa = 6.85NIGSITSAMRR306 pKa = 11.84NMSQWISATFPASKK320 pKa = 9.33TLRR323 pKa = 11.84HH324 pKa = 5.27YY325 pKa = 8.9MQAGSHH331 pKa = 5.01EE332 pKa = 4.17TSNYY336 pKa = 9.7LFAEE340 pKa = 4.29IALPTWNTTQITITVPVSQTNSDD363 pKa = 3.47KK364 pKa = 10.79FLPTAAPDD372 pKa = 3.5QNATEE377 pKa = 4.41LAHH380 pKa = 7.46DD381 pKa = 4.24SGFLTVPTAPADD393 pKa = 3.54TDD395 pKa = 3.96VNGTSVWNIAAEE407 pKa = 4.44RR408 pKa = 11.84AASTPPANPMYY419 pKa = 10.92ASLRR423 pKa = 11.84SSATRR428 pKa = 11.84PDD430 pKa = 4.22PYY432 pKa = 10.93VNPIPHH438 pKa = 5.48VTFDD442 pKa = 3.81VNLHH446 pKa = 5.19VLPRR450 pKa = 11.84MYY452 pKa = 10.18IYY454 pKa = 10.88APYY457 pKa = 8.5VTNTGALGSVITSGKK472 pKa = 10.0LIEE475 pKa = 5.05SGDD478 pKa = 3.4ISGIMQIVPSTTTPLLYY495 pKa = 10.6EE496 pKa = 4.04NSQFHH501 pKa = 7.33DD502 pKa = 3.86GAIRR506 pKa = 11.84LSRR509 pKa = 11.84TRR511 pKa = 11.84SALFHH516 pKa = 6.84AGIQIHH522 pKa = 4.94MQEE525 pKa = 3.99RR526 pKa = 11.84PARR529 pKa = 11.84RR530 pKa = 11.84TLDD533 pKa = 3.21APIAFFRR540 pKa = 11.84GLTSGLRR547 pKa = 11.84YY548 pKa = 8.53PVPSTGPVYY557 pKa = 10.73APSAFPNSSQARR569 pKa = 11.84LLPGARR575 pKa = 11.84WTSHH579 pKa = 4.42ATNFFHH585 pKa = 6.7TLNVFGRR592 pKa = 11.84DD593 pKa = 3.54LSHH596 pKa = 7.8DD597 pKa = 4.08LQLHH601 pKa = 6.37ADD603 pKa = 3.4SSFNIWSGLRR613 pKa = 11.84FRR615 pKa = 11.84YY616 pKa = 6.75MTNTGPVVYY625 pKa = 9.4VLPSARR631 pKa = 11.84HH632 pKa = 4.68IFGARR637 pKa = 11.84ARR639 pKa = 11.84SYY641 pKa = 9.58GTEE644 pKa = 3.97HH645 pKa = 7.07PALRR649 pKa = 11.84LPLL652 pKa = 4.15
Molecular weight: 71.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1405 |
652 |
753 |
702.5 |
79.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.616 ± 2.646 | 0.712 ± 0.485 |
5.338 ± 0.71 | 3.559 ± 1.378 |
6.69 ± 0.9 | 3.701 ± 0.508 |
3.63 ± 0.278 | 6.335 ± 0.553 |
2.278 ± 1.237 | 9.68 ± 0.325 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.349 ± 0.176 | 5.48 ± 1.072 |
7.829 ± 0.1 | 3.203 ± 0.117 |
5.836 ± 0.423 | 8.541 ± 0.868 |
7.046 ± 0.737 | 4.769 ± 0.408 |
1.21 ± 0.012 | 4.199 ± 0.353 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
