
Shahe picorna-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
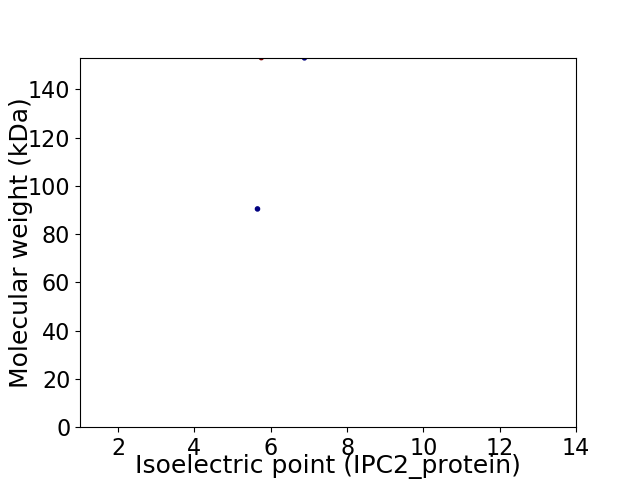
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJ24|A0A1L3KJ24_9VIRU SF3 helicase domain-containing protein OS=Shahe picorna-like virus 12 OX=1923442 PE=4 SV=1
MM1 pKa = 7.6NYY3 pKa = 10.49DD4 pKa = 3.51QFLHH8 pKa = 6.55KK9 pKa = 10.21PLFIDD14 pKa = 3.33NFNWSTVDD22 pKa = 3.13AAGINIGGVQIPRR35 pKa = 11.84DD36 pKa = 3.48IFNTNSFLAIPWKK49 pKa = 10.02SSCYY53 pKa = 9.88YY54 pKa = 9.76RR55 pKa = 11.84LKK57 pKa = 10.74GKK59 pKa = 10.13LVIQVAGTVRR69 pKa = 11.84HH70 pKa = 5.85SGILLASALPFVAPTNFPLNSMLAVPHH97 pKa = 6.31AFVYY101 pKa = 10.82ANQSNPAIVEE111 pKa = 3.95IPFYY115 pKa = 10.79NPSPLRR121 pKa = 11.84QTQDD125 pKa = 2.6NDD127 pKa = 3.98RR128 pKa = 11.84TCHH131 pKa = 6.7LAIQRR136 pKa = 11.84DD137 pKa = 4.01GSDD140 pKa = 3.09SYY142 pKa = 11.86SDD144 pKa = 3.37VFIRR148 pKa = 11.84IINPLSAPTGGSTTVVVTVHH168 pKa = 6.54MILDD172 pKa = 3.66EE173 pKa = 4.65LEE175 pKa = 4.81FYY177 pKa = 10.54TPAPVDD183 pKa = 3.16IVYY186 pKa = 9.99AQSKK190 pKa = 10.41VITQAMDD197 pKa = 3.29SVASILKK204 pKa = 9.85QKK206 pKa = 10.76SSDD209 pKa = 3.7YY210 pKa = 10.39IDD212 pKa = 3.61NVRR215 pKa = 11.84GALHH219 pKa = 5.79QWTGLHH225 pKa = 6.12NPNIDD230 pKa = 3.37APLNKK235 pKa = 9.95HH236 pKa = 6.33FMQTRR241 pKa = 11.84ANPNVIDD248 pKa = 4.68AVTTYY253 pKa = 8.96DD254 pKa = 3.39TLHH257 pKa = 7.55PYY259 pKa = 10.15TMDD262 pKa = 3.29EE263 pKa = 4.35SGTTSAAFFHH273 pKa = 6.59TEE275 pKa = 3.61TDD277 pKa = 3.61EE278 pKa = 4.33MDD280 pKa = 3.3MKK282 pKa = 11.0YY283 pKa = 10.51LLSIPQVISTRR294 pKa = 11.84IPIPLNSAVGSQLFCRR310 pKa = 11.84PITPFMGVKK319 pKa = 10.48NSVTDD324 pKa = 3.18LHH326 pKa = 6.45TIKK329 pKa = 10.67SIQSKK334 pKa = 10.05LAFCASHH341 pKa = 6.63WSGDD345 pKa = 3.84MEE347 pKa = 6.1LMLQVDD353 pKa = 4.05MSNTQFLKK361 pKa = 11.04LLVVLDD367 pKa = 3.46YY368 pKa = 10.8TRR370 pKa = 11.84NINSTNLGIAGSPASFSPYY389 pKa = 10.12QGTLTHH395 pKa = 6.05TLEE398 pKa = 4.29FAGGGSIQCVTLPFMSAFRR417 pKa = 11.84QLPLTTDD424 pKa = 2.51WRR426 pKa = 11.84ANALSHH432 pKa = 5.95GLVRR436 pKa = 11.84IYY438 pKa = 10.78VLQPPVVSDD447 pKa = 3.81DD448 pKa = 3.73VAVPEE453 pKa = 4.23LNVYY457 pKa = 7.81YY458 pKa = 10.27RR459 pKa = 11.84CKK461 pKa = 10.92EE462 pKa = 3.73NFQLYY467 pKa = 8.85GFNYY471 pKa = 9.71RR472 pKa = 11.84RR473 pKa = 11.84GHH475 pKa = 5.75YY476 pKa = 9.87SATSPIPPIAAMGEE490 pKa = 4.16EE491 pKa = 4.68VLDD494 pKa = 4.06SMTTQSMAMSDD505 pKa = 3.44SSMLDD510 pKa = 3.01TTTSQEE516 pKa = 4.22CVTDD520 pKa = 3.89KK521 pKa = 11.27GPSKK525 pKa = 10.39PSVVYY530 pKa = 10.62GPGNLRR536 pKa = 11.84PIVSVRR542 pKa = 11.84DD543 pKa = 3.8IIRR546 pKa = 11.84RR547 pKa = 11.84VYY549 pKa = 9.64PVYY552 pKa = 11.2SNVLTPSVTSNVNYY566 pKa = 10.65LNLPLQGLFRR576 pKa = 11.84SRR578 pKa = 11.84QIGTNFSGTASDD590 pKa = 3.98ATPLSIISDD599 pKa = 3.49MFYY602 pKa = 11.13GFRR605 pKa = 11.84GSMKK609 pKa = 10.16LKK611 pKa = 10.7IMITGANASAVHH623 pKa = 6.56FYY625 pKa = 10.14PPNVSVTSEE634 pKa = 4.14GVNKK638 pKa = 9.64VYY640 pKa = 10.7TMTTPSTTNTTFIADD655 pKa = 3.9EE656 pKa = 4.39VGSCLPFSDD665 pKa = 4.6SFRR668 pKa = 11.84MIPGTTYY675 pKa = 11.02QEE677 pKa = 3.77MSDD680 pKa = 3.6FNRR683 pKa = 11.84RR684 pKa = 11.84PSIVHH689 pKa = 4.84TTTGVGSLADD699 pKa = 3.46SVSIHH704 pKa = 7.29DD705 pKa = 3.21IHH707 pKa = 8.23VPYY710 pKa = 9.91MSHH713 pKa = 6.85QDD715 pKa = 3.37FVTNDD720 pKa = 3.2GKK722 pKa = 11.28LLGLNTPQSSSDD734 pKa = 3.3SLGNLKK740 pKa = 9.97ISYY743 pKa = 8.78TPVIVSPAGATVVLSRR759 pKa = 11.84VHH761 pKa = 4.74ITIFVGLGDD770 pKa = 3.87DD771 pKa = 4.04ARR773 pKa = 11.84FGMLINARR781 pKa = 11.84EE782 pKa = 4.09LNPYY786 pKa = 9.24FALDD790 pKa = 3.74GVTKK794 pKa = 10.41IQLDD798 pKa = 4.74PIQHH802 pKa = 6.31ANVVVGNTTVPSLISDD818 pKa = 3.94SGAAQAYY825 pKa = 9.45IGG827 pKa = 3.94
MM1 pKa = 7.6NYY3 pKa = 10.49DD4 pKa = 3.51QFLHH8 pKa = 6.55KK9 pKa = 10.21PLFIDD14 pKa = 3.33NFNWSTVDD22 pKa = 3.13AAGINIGGVQIPRR35 pKa = 11.84DD36 pKa = 3.48IFNTNSFLAIPWKK49 pKa = 10.02SSCYY53 pKa = 9.88YY54 pKa = 9.76RR55 pKa = 11.84LKK57 pKa = 10.74GKK59 pKa = 10.13LVIQVAGTVRR69 pKa = 11.84HH70 pKa = 5.85SGILLASALPFVAPTNFPLNSMLAVPHH97 pKa = 6.31AFVYY101 pKa = 10.82ANQSNPAIVEE111 pKa = 3.95IPFYY115 pKa = 10.79NPSPLRR121 pKa = 11.84QTQDD125 pKa = 2.6NDD127 pKa = 3.98RR128 pKa = 11.84TCHH131 pKa = 6.7LAIQRR136 pKa = 11.84DD137 pKa = 4.01GSDD140 pKa = 3.09SYY142 pKa = 11.86SDD144 pKa = 3.37VFIRR148 pKa = 11.84IINPLSAPTGGSTTVVVTVHH168 pKa = 6.54MILDD172 pKa = 3.66EE173 pKa = 4.65LEE175 pKa = 4.81FYY177 pKa = 10.54TPAPVDD183 pKa = 3.16IVYY186 pKa = 9.99AQSKK190 pKa = 10.41VITQAMDD197 pKa = 3.29SVASILKK204 pKa = 9.85QKK206 pKa = 10.76SSDD209 pKa = 3.7YY210 pKa = 10.39IDD212 pKa = 3.61NVRR215 pKa = 11.84GALHH219 pKa = 5.79QWTGLHH225 pKa = 6.12NPNIDD230 pKa = 3.37APLNKK235 pKa = 9.95HH236 pKa = 6.33FMQTRR241 pKa = 11.84ANPNVIDD248 pKa = 4.68AVTTYY253 pKa = 8.96DD254 pKa = 3.39TLHH257 pKa = 7.55PYY259 pKa = 10.15TMDD262 pKa = 3.29EE263 pKa = 4.35SGTTSAAFFHH273 pKa = 6.59TEE275 pKa = 3.61TDD277 pKa = 3.61EE278 pKa = 4.33MDD280 pKa = 3.3MKK282 pKa = 11.0YY283 pKa = 10.51LLSIPQVISTRR294 pKa = 11.84IPIPLNSAVGSQLFCRR310 pKa = 11.84PITPFMGVKK319 pKa = 10.48NSVTDD324 pKa = 3.18LHH326 pKa = 6.45TIKK329 pKa = 10.67SIQSKK334 pKa = 10.05LAFCASHH341 pKa = 6.63WSGDD345 pKa = 3.84MEE347 pKa = 6.1LMLQVDD353 pKa = 4.05MSNTQFLKK361 pKa = 11.04LLVVLDD367 pKa = 3.46YY368 pKa = 10.8TRR370 pKa = 11.84NINSTNLGIAGSPASFSPYY389 pKa = 10.12QGTLTHH395 pKa = 6.05TLEE398 pKa = 4.29FAGGGSIQCVTLPFMSAFRR417 pKa = 11.84QLPLTTDD424 pKa = 2.51WRR426 pKa = 11.84ANALSHH432 pKa = 5.95GLVRR436 pKa = 11.84IYY438 pKa = 10.78VLQPPVVSDD447 pKa = 3.81DD448 pKa = 3.73VAVPEE453 pKa = 4.23LNVYY457 pKa = 7.81YY458 pKa = 10.27RR459 pKa = 11.84CKK461 pKa = 10.92EE462 pKa = 3.73NFQLYY467 pKa = 8.85GFNYY471 pKa = 9.71RR472 pKa = 11.84RR473 pKa = 11.84GHH475 pKa = 5.75YY476 pKa = 9.87SATSPIPPIAAMGEE490 pKa = 4.16EE491 pKa = 4.68VLDD494 pKa = 4.06SMTTQSMAMSDD505 pKa = 3.44SSMLDD510 pKa = 3.01TTTSQEE516 pKa = 4.22CVTDD520 pKa = 3.89KK521 pKa = 11.27GPSKK525 pKa = 10.39PSVVYY530 pKa = 10.62GPGNLRR536 pKa = 11.84PIVSVRR542 pKa = 11.84DD543 pKa = 3.8IIRR546 pKa = 11.84RR547 pKa = 11.84VYY549 pKa = 9.64PVYY552 pKa = 11.2SNVLTPSVTSNVNYY566 pKa = 10.65LNLPLQGLFRR576 pKa = 11.84SRR578 pKa = 11.84QIGTNFSGTASDD590 pKa = 3.98ATPLSIISDD599 pKa = 3.49MFYY602 pKa = 11.13GFRR605 pKa = 11.84GSMKK609 pKa = 10.16LKK611 pKa = 10.7IMITGANASAVHH623 pKa = 6.56FYY625 pKa = 10.14PPNVSVTSEE634 pKa = 4.14GVNKK638 pKa = 9.64VYY640 pKa = 10.7TMTTPSTTNTTFIADD655 pKa = 3.9EE656 pKa = 4.39VGSCLPFSDD665 pKa = 4.6SFRR668 pKa = 11.84MIPGTTYY675 pKa = 11.02QEE677 pKa = 3.77MSDD680 pKa = 3.6FNRR683 pKa = 11.84RR684 pKa = 11.84PSIVHH689 pKa = 4.84TTTGVGSLADD699 pKa = 3.46SVSIHH704 pKa = 7.29DD705 pKa = 3.21IHH707 pKa = 8.23VPYY710 pKa = 9.91MSHH713 pKa = 6.85QDD715 pKa = 3.37FVTNDD720 pKa = 3.2GKK722 pKa = 11.28LLGLNTPQSSSDD734 pKa = 3.3SLGNLKK740 pKa = 9.97ISYY743 pKa = 8.78TPVIVSPAGATVVLSRR759 pKa = 11.84VHH761 pKa = 4.74ITIFVGLGDD770 pKa = 3.87DD771 pKa = 4.04ARR773 pKa = 11.84FGMLINARR781 pKa = 11.84EE782 pKa = 4.09LNPYY786 pKa = 9.24FALDD790 pKa = 3.74GVTKK794 pKa = 10.41IQLDD798 pKa = 4.74PIQHH802 pKa = 6.31ANVVVGNTTVPSLISDD818 pKa = 3.94SGAAQAYY825 pKa = 9.45IGG827 pKa = 3.94
Molecular weight: 90.43 kDa
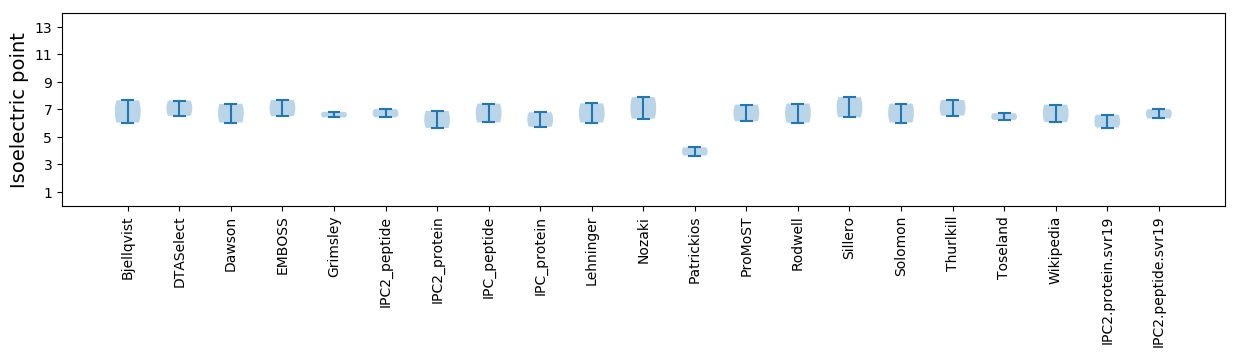
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJ24|A0A1L3KJ24_9VIRU SF3 helicase domain-containing protein OS=Shahe picorna-like virus 12 OX=1923442 PE=4 SV=1
MM1 pKa = 7.46VYY3 pKa = 10.61NSSTNQGDD11 pKa = 3.74RR12 pKa = 11.84KK13 pKa = 10.6NGNKK17 pKa = 9.73NFLFINNCFSNYY29 pKa = 9.93FIMANPYY36 pKa = 9.26RR37 pKa = 11.84CVRR40 pKa = 11.84SSKK43 pKa = 10.68NAFSSNEE50 pKa = 3.91WPVLKK55 pKa = 10.68CGLLLVKK62 pKa = 9.29FNPAAGSRR70 pKa = 11.84ILYY73 pKa = 10.04RR74 pKa = 11.84EE75 pKa = 3.87TFIQSVLIHH84 pKa = 5.99VDD86 pKa = 3.02RR87 pKa = 11.84MRR89 pKa = 11.84VQGLTDD95 pKa = 4.58IISDD99 pKa = 3.67MKK101 pKa = 10.57EE102 pKa = 3.7LLVSVFGVFRR112 pKa = 11.84TMVKK116 pKa = 10.15VVQKK120 pKa = 10.08CSNMKK125 pKa = 9.04TATALMDD132 pKa = 3.67WLLDD136 pKa = 4.6AILLLSDD143 pKa = 3.19VLTRR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 10.17VYY151 pKa = 10.32GMNIIITLLRR161 pKa = 11.84VFKK164 pKa = 10.43LYY166 pKa = 10.45KK167 pKa = 10.26DD168 pKa = 4.18SPLHH172 pKa = 4.89PQSWQGLLVSLVSFGVPAAVIDD194 pKa = 3.93VVKK197 pKa = 10.78RR198 pKa = 11.84MSLLSNTKK206 pKa = 10.09VLDD209 pKa = 4.21DD210 pKa = 4.5FSLFHH215 pKa = 6.4QLISVCMDD223 pKa = 3.82TLASFIEE230 pKa = 4.46TLGFTQTPVVKK241 pKa = 10.41QIKK244 pKa = 9.9EE245 pKa = 3.9YY246 pKa = 10.86GKK248 pKa = 10.45DD249 pKa = 3.63LIQHH253 pKa = 6.64HH254 pKa = 6.54SLLADD259 pKa = 3.29IKK261 pKa = 11.35DD262 pKa = 3.39MCKK265 pKa = 9.96LVKK268 pKa = 9.5KK269 pKa = 10.24DD270 pKa = 3.35RR271 pKa = 11.84RR272 pKa = 11.84IFSEE276 pKa = 3.79SHH278 pKa = 4.68FRR280 pKa = 11.84EE281 pKa = 4.65KK282 pKa = 10.6IFTTHH287 pKa = 5.94EE288 pKa = 4.18KK289 pKa = 10.8VVGNTILLDD298 pKa = 3.46WGKK301 pKa = 8.73TSPSVANEE309 pKa = 3.12IATIKK314 pKa = 10.2EE315 pKa = 4.16LNKK318 pKa = 10.39LALAYY323 pKa = 9.28EE324 pKa = 4.33NNSRR328 pKa = 11.84QEE330 pKa = 4.02PLCMCFEE337 pKa = 4.57GPPGVFKK344 pKa = 10.49STIMSLLLKK353 pKa = 10.36ALNKK357 pKa = 9.97SVYY360 pKa = 8.41SHH362 pKa = 5.5QVKK365 pKa = 10.72SLMDD369 pKa = 3.69GKK371 pKa = 10.44DD372 pKa = 3.51FYY374 pKa = 11.56DD375 pKa = 3.62QYY377 pKa = 11.83DD378 pKa = 3.65NQDD381 pKa = 3.0IMLMDD386 pKa = 4.86DD387 pKa = 4.51LGAGGDD393 pKa = 3.63SQFRR397 pKa = 11.84FLVNLISPIKK407 pKa = 10.48YY408 pKa = 9.72PLDD411 pKa = 3.92CASEE415 pKa = 4.16KK416 pKa = 11.19LKK418 pKa = 9.8DD419 pKa = 3.55TKK421 pKa = 10.76FFTSEE426 pKa = 3.62AVLFTTNKK434 pKa = 9.56FSTIRR439 pKa = 11.84STTSGSGIVDD449 pKa = 4.0LNAVKK454 pKa = 10.41RR455 pKa = 11.84RR456 pKa = 11.84VVLIDD461 pKa = 3.88FSHH464 pKa = 6.64VKK466 pKa = 10.44RR467 pKa = 11.84KK468 pKa = 10.19GDD470 pKa = 3.59WIEE473 pKa = 4.88GILKK477 pKa = 10.37ASEE480 pKa = 4.05FSLEE484 pKa = 4.03HH485 pKa = 6.62KK486 pKa = 10.34EE487 pKa = 3.93YY488 pKa = 11.25VPLFSIDD495 pKa = 3.29ITPEE499 pKa = 3.72CDD501 pKa = 2.78VGNKK505 pKa = 9.06VKK507 pKa = 10.86LLGWLSAIYY516 pKa = 10.23EE517 pKa = 3.96MSIDD521 pKa = 3.73LKK523 pKa = 10.88KK524 pKa = 10.98SHH526 pKa = 6.51VNNNAANEE534 pKa = 4.1DD535 pKa = 3.89LLQEE539 pKa = 3.94IRR541 pKa = 11.84KK542 pKa = 8.22ATSEE546 pKa = 4.26WKK548 pKa = 9.23TKK550 pKa = 10.11AQSAYY555 pKa = 10.63DD556 pKa = 3.92PYY558 pKa = 10.23TGVGILQSYY567 pKa = 6.43MHH569 pKa = 7.03HH570 pKa = 6.9AFAICSSFVEE580 pKa = 4.41EE581 pKa = 3.89RR582 pKa = 11.84ASYY585 pKa = 9.79FSSYY589 pKa = 10.43APPVRR594 pKa = 11.84TMIGNFVSKK603 pKa = 10.83VEE605 pKa = 4.17EE606 pKa = 4.2VSSDD610 pKa = 3.53MIVWGSIIGISSILLSYY627 pKa = 11.42GLFTYY632 pKa = 9.19FFSQEE637 pKa = 3.61QDD639 pKa = 2.78EE640 pKa = 4.46MQLQKK645 pKa = 10.87EE646 pKa = 3.95QDD648 pKa = 3.02IRR650 pKa = 11.84CQVVQGSTMAEE661 pKa = 3.57AVAKK665 pKa = 8.53QTFFMEE671 pKa = 5.4FIKK674 pKa = 10.74KK675 pKa = 10.24DD676 pKa = 3.42GSKK679 pKa = 10.6EE680 pKa = 3.96KK681 pKa = 10.31ATGTVSGHH689 pKa = 6.69FILAPWHH696 pKa = 6.01AAKK699 pKa = 10.58DD700 pKa = 4.0CIRR703 pKa = 11.84ISILSKK709 pKa = 10.88VADD712 pKa = 4.1CRR714 pKa = 11.84LLDD717 pKa = 3.91NAPCTKK723 pKa = 10.24VYY725 pKa = 10.05EE726 pKa = 4.67AYY728 pKa = 10.75SSDD731 pKa = 3.67LVVLKK736 pKa = 10.55IPEE739 pKa = 4.11TLIVPFKK746 pKa = 11.06SLVKK750 pKa = 10.75VFDD753 pKa = 4.19YY754 pKa = 11.01KK755 pKa = 11.03CDD757 pKa = 3.54ALKK760 pKa = 10.86GHH762 pKa = 6.2TLITPYY768 pKa = 11.07GCLPVVKK775 pKa = 9.03RR776 pKa = 11.84TAGCSILNIYY786 pKa = 9.29GKK788 pKa = 9.83NFQLDD793 pKa = 3.77DD794 pKa = 4.51SNSFPYY800 pKa = 9.21TYY802 pKa = 11.33SMDD805 pKa = 3.9GLCGSLLFDD814 pKa = 4.1NATGVIGAHH823 pKa = 5.91IAGNEE828 pKa = 3.94QYY830 pKa = 11.11GRR832 pKa = 11.84ALKK835 pKa = 10.19WNTGVFSEE843 pKa = 5.2VMSLLKK849 pKa = 10.39GDD851 pKa = 4.06RR852 pKa = 11.84FLLEE856 pKa = 5.65LDD858 pKa = 4.07FKK860 pKa = 11.85DD861 pKa = 3.69NGKK864 pKa = 10.24LSSGTQLDD872 pKa = 4.4TNFHH876 pKa = 5.85QSTPKK881 pKa = 8.12VTKK884 pKa = 10.03IEE886 pKa = 3.99PTIFSHH892 pKa = 6.47MFPKK896 pKa = 10.43HH897 pKa = 6.13KK898 pKa = 10.7EE899 pKa = 3.76PVNLSKK905 pKa = 11.12YY906 pKa = 8.58GQHH909 pKa = 5.34TVKK912 pKa = 10.77DD913 pKa = 3.83VAKK916 pKa = 10.5KK917 pKa = 10.37SFSKK921 pKa = 10.73VKK923 pKa = 10.45EE924 pKa = 3.86ITQVEE929 pKa = 4.55LDD931 pKa = 3.74FGLDD935 pKa = 3.57YY936 pKa = 11.09LSTLMQPFSSLSEE949 pKa = 3.79QDD951 pKa = 3.34TVKK954 pKa = 10.95GFDD957 pKa = 3.24KK958 pKa = 10.67VAKK961 pKa = 10.22INMDD965 pKa = 2.85KK966 pKa = 10.08STGIGCEE973 pKa = 3.85RR974 pKa = 11.84DD975 pKa = 2.88RR976 pKa = 11.84AAYY979 pKa = 9.61INFEE983 pKa = 4.08RR984 pKa = 11.84GYY986 pKa = 8.85YY987 pKa = 8.05TQLLRR992 pKa = 11.84KK993 pKa = 8.78EE994 pKa = 4.23IKK996 pKa = 10.4DD997 pKa = 3.69IEE999 pKa = 4.76DD1000 pKa = 3.44NLLKK1004 pKa = 10.77DD1005 pKa = 3.29EE1006 pKa = 4.65HH1007 pKa = 7.45VNLNLWVAKK1016 pKa = 7.78EE1017 pKa = 3.97TLKK1020 pKa = 11.36DD1021 pKa = 3.6EE1022 pKa = 4.3VRR1024 pKa = 11.84SVSKK1028 pKa = 10.64DD1029 pKa = 3.13QEE1031 pKa = 4.03PRR1033 pKa = 11.84SFRR1036 pKa = 11.84VLRR1039 pKa = 11.84FPINVLCKK1047 pKa = 9.8QLTGEE1052 pKa = 4.2MVNNIIANKK1061 pKa = 8.93FSNGIMIGINPYY1073 pKa = 9.43SEE1075 pKa = 4.0WEE1077 pKa = 4.01TLYY1080 pKa = 9.89NTVGYY1085 pKa = 8.86KK1086 pKa = 10.27AKK1088 pKa = 10.75GIIAADD1094 pKa = 3.18IKK1096 pKa = 11.17KK1097 pKa = 10.12FDD1099 pKa = 4.01GGMLPQVQHH1108 pKa = 5.93GVVDD1112 pKa = 4.81TILSFFSGTYY1122 pKa = 9.98KK1123 pKa = 9.71EE1124 pKa = 4.46TEE1126 pKa = 3.95LLKK1129 pKa = 11.15GILHH1133 pKa = 6.14NFINNPVAVNDD1144 pKa = 3.89DD1145 pKa = 4.05VYY1147 pKa = 10.82LTTHH1151 pKa = 6.38SMPSGCYY1158 pKa = 8.15LTAIMNSMVQKK1169 pKa = 10.57VYY1171 pKa = 9.16TAMWYY1176 pKa = 10.14KK1177 pKa = 10.51HH1178 pKa = 6.37VIPDD1182 pKa = 3.64ATVTQYY1188 pKa = 11.34KK1189 pKa = 9.73QDD1191 pKa = 3.27VYY1193 pKa = 11.42DD1194 pKa = 4.46AVYY1197 pKa = 10.93GDD1199 pKa = 5.78DD1200 pKa = 4.19KK1201 pKa = 11.53LCSIKK1206 pKa = 10.89NKK1208 pKa = 10.16EE1209 pKa = 4.63DD1210 pKa = 3.13ILNAITMRR1218 pKa = 11.84DD1219 pKa = 3.59YY1220 pKa = 11.13FQLIGLDD1227 pKa = 4.07LSTADD1232 pKa = 4.35KK1233 pKa = 11.38KK1234 pKa = 11.23EE1235 pKa = 3.54ITQPFDD1241 pKa = 2.5SWEE1244 pKa = 3.99DD1245 pKa = 3.43VNFLKK1250 pKa = 10.61RR1251 pKa = 11.84KK1252 pKa = 9.0FVFHH1256 pKa = 6.8PVLKK1260 pKa = 10.35RR1261 pKa = 11.84IMCPLIDD1268 pKa = 3.51EE1269 pKa = 4.97TFYY1272 pKa = 12.01SMLAWWDD1279 pKa = 3.44TSKK1282 pKa = 11.37DD1283 pKa = 3.76FNDD1286 pKa = 4.15VIDD1289 pKa = 4.22GKK1291 pKa = 10.8LRR1293 pKa = 11.84SFTYY1297 pKa = 10.22EE1298 pKa = 3.51SYY1300 pKa = 10.98LRR1302 pKa = 11.84EE1303 pKa = 4.53DD1304 pKa = 3.31NDD1306 pKa = 4.32SNRR1309 pKa = 11.84EE1310 pKa = 3.88KK1311 pKa = 11.12LKK1313 pKa = 10.39MYY1315 pKa = 10.34LEE1317 pKa = 4.71NVAGMTWIMPSNMEE1331 pKa = 4.44MYY1333 pKa = 10.49RR1334 pKa = 11.84QFLKK1338 pKa = 11.3NEE1340 pKa = 3.71VDD1342 pKa = 3.57YY1343 pKa = 8.4TTYY1346 pKa = 11.43AA1347 pKa = 3.21
MM1 pKa = 7.46VYY3 pKa = 10.61NSSTNQGDD11 pKa = 3.74RR12 pKa = 11.84KK13 pKa = 10.6NGNKK17 pKa = 9.73NFLFINNCFSNYY29 pKa = 9.93FIMANPYY36 pKa = 9.26RR37 pKa = 11.84CVRR40 pKa = 11.84SSKK43 pKa = 10.68NAFSSNEE50 pKa = 3.91WPVLKK55 pKa = 10.68CGLLLVKK62 pKa = 9.29FNPAAGSRR70 pKa = 11.84ILYY73 pKa = 10.04RR74 pKa = 11.84EE75 pKa = 3.87TFIQSVLIHH84 pKa = 5.99VDD86 pKa = 3.02RR87 pKa = 11.84MRR89 pKa = 11.84VQGLTDD95 pKa = 4.58IISDD99 pKa = 3.67MKK101 pKa = 10.57EE102 pKa = 3.7LLVSVFGVFRR112 pKa = 11.84TMVKK116 pKa = 10.15VVQKK120 pKa = 10.08CSNMKK125 pKa = 9.04TATALMDD132 pKa = 3.67WLLDD136 pKa = 4.6AILLLSDD143 pKa = 3.19VLTRR147 pKa = 11.84RR148 pKa = 11.84KK149 pKa = 10.17VYY151 pKa = 10.32GMNIIITLLRR161 pKa = 11.84VFKK164 pKa = 10.43LYY166 pKa = 10.45KK167 pKa = 10.26DD168 pKa = 4.18SPLHH172 pKa = 4.89PQSWQGLLVSLVSFGVPAAVIDD194 pKa = 3.93VVKK197 pKa = 10.78RR198 pKa = 11.84MSLLSNTKK206 pKa = 10.09VLDD209 pKa = 4.21DD210 pKa = 4.5FSLFHH215 pKa = 6.4QLISVCMDD223 pKa = 3.82TLASFIEE230 pKa = 4.46TLGFTQTPVVKK241 pKa = 10.41QIKK244 pKa = 9.9EE245 pKa = 3.9YY246 pKa = 10.86GKK248 pKa = 10.45DD249 pKa = 3.63LIQHH253 pKa = 6.64HH254 pKa = 6.54SLLADD259 pKa = 3.29IKK261 pKa = 11.35DD262 pKa = 3.39MCKK265 pKa = 9.96LVKK268 pKa = 9.5KK269 pKa = 10.24DD270 pKa = 3.35RR271 pKa = 11.84RR272 pKa = 11.84IFSEE276 pKa = 3.79SHH278 pKa = 4.68FRR280 pKa = 11.84EE281 pKa = 4.65KK282 pKa = 10.6IFTTHH287 pKa = 5.94EE288 pKa = 4.18KK289 pKa = 10.8VVGNTILLDD298 pKa = 3.46WGKK301 pKa = 8.73TSPSVANEE309 pKa = 3.12IATIKK314 pKa = 10.2EE315 pKa = 4.16LNKK318 pKa = 10.39LALAYY323 pKa = 9.28EE324 pKa = 4.33NNSRR328 pKa = 11.84QEE330 pKa = 4.02PLCMCFEE337 pKa = 4.57GPPGVFKK344 pKa = 10.49STIMSLLLKK353 pKa = 10.36ALNKK357 pKa = 9.97SVYY360 pKa = 8.41SHH362 pKa = 5.5QVKK365 pKa = 10.72SLMDD369 pKa = 3.69GKK371 pKa = 10.44DD372 pKa = 3.51FYY374 pKa = 11.56DD375 pKa = 3.62QYY377 pKa = 11.83DD378 pKa = 3.65NQDD381 pKa = 3.0IMLMDD386 pKa = 4.86DD387 pKa = 4.51LGAGGDD393 pKa = 3.63SQFRR397 pKa = 11.84FLVNLISPIKK407 pKa = 10.48YY408 pKa = 9.72PLDD411 pKa = 3.92CASEE415 pKa = 4.16KK416 pKa = 11.19LKK418 pKa = 9.8DD419 pKa = 3.55TKK421 pKa = 10.76FFTSEE426 pKa = 3.62AVLFTTNKK434 pKa = 9.56FSTIRR439 pKa = 11.84STTSGSGIVDD449 pKa = 4.0LNAVKK454 pKa = 10.41RR455 pKa = 11.84RR456 pKa = 11.84VVLIDD461 pKa = 3.88FSHH464 pKa = 6.64VKK466 pKa = 10.44RR467 pKa = 11.84KK468 pKa = 10.19GDD470 pKa = 3.59WIEE473 pKa = 4.88GILKK477 pKa = 10.37ASEE480 pKa = 4.05FSLEE484 pKa = 4.03HH485 pKa = 6.62KK486 pKa = 10.34EE487 pKa = 3.93YY488 pKa = 11.25VPLFSIDD495 pKa = 3.29ITPEE499 pKa = 3.72CDD501 pKa = 2.78VGNKK505 pKa = 9.06VKK507 pKa = 10.86LLGWLSAIYY516 pKa = 10.23EE517 pKa = 3.96MSIDD521 pKa = 3.73LKK523 pKa = 10.88KK524 pKa = 10.98SHH526 pKa = 6.51VNNNAANEE534 pKa = 4.1DD535 pKa = 3.89LLQEE539 pKa = 3.94IRR541 pKa = 11.84KK542 pKa = 8.22ATSEE546 pKa = 4.26WKK548 pKa = 9.23TKK550 pKa = 10.11AQSAYY555 pKa = 10.63DD556 pKa = 3.92PYY558 pKa = 10.23TGVGILQSYY567 pKa = 6.43MHH569 pKa = 7.03HH570 pKa = 6.9AFAICSSFVEE580 pKa = 4.41EE581 pKa = 3.89RR582 pKa = 11.84ASYY585 pKa = 9.79FSSYY589 pKa = 10.43APPVRR594 pKa = 11.84TMIGNFVSKK603 pKa = 10.83VEE605 pKa = 4.17EE606 pKa = 4.2VSSDD610 pKa = 3.53MIVWGSIIGISSILLSYY627 pKa = 11.42GLFTYY632 pKa = 9.19FFSQEE637 pKa = 3.61QDD639 pKa = 2.78EE640 pKa = 4.46MQLQKK645 pKa = 10.87EE646 pKa = 3.95QDD648 pKa = 3.02IRR650 pKa = 11.84CQVVQGSTMAEE661 pKa = 3.57AVAKK665 pKa = 8.53QTFFMEE671 pKa = 5.4FIKK674 pKa = 10.74KK675 pKa = 10.24DD676 pKa = 3.42GSKK679 pKa = 10.6EE680 pKa = 3.96KK681 pKa = 10.31ATGTVSGHH689 pKa = 6.69FILAPWHH696 pKa = 6.01AAKK699 pKa = 10.58DD700 pKa = 4.0CIRR703 pKa = 11.84ISILSKK709 pKa = 10.88VADD712 pKa = 4.1CRR714 pKa = 11.84LLDD717 pKa = 3.91NAPCTKK723 pKa = 10.24VYY725 pKa = 10.05EE726 pKa = 4.67AYY728 pKa = 10.75SSDD731 pKa = 3.67LVVLKK736 pKa = 10.55IPEE739 pKa = 4.11TLIVPFKK746 pKa = 11.06SLVKK750 pKa = 10.75VFDD753 pKa = 4.19YY754 pKa = 11.01KK755 pKa = 11.03CDD757 pKa = 3.54ALKK760 pKa = 10.86GHH762 pKa = 6.2TLITPYY768 pKa = 11.07GCLPVVKK775 pKa = 9.03RR776 pKa = 11.84TAGCSILNIYY786 pKa = 9.29GKK788 pKa = 9.83NFQLDD793 pKa = 3.77DD794 pKa = 4.51SNSFPYY800 pKa = 9.21TYY802 pKa = 11.33SMDD805 pKa = 3.9GLCGSLLFDD814 pKa = 4.1NATGVIGAHH823 pKa = 5.91IAGNEE828 pKa = 3.94QYY830 pKa = 11.11GRR832 pKa = 11.84ALKK835 pKa = 10.19WNTGVFSEE843 pKa = 5.2VMSLLKK849 pKa = 10.39GDD851 pKa = 4.06RR852 pKa = 11.84FLLEE856 pKa = 5.65LDD858 pKa = 4.07FKK860 pKa = 11.85DD861 pKa = 3.69NGKK864 pKa = 10.24LSSGTQLDD872 pKa = 4.4TNFHH876 pKa = 5.85QSTPKK881 pKa = 8.12VTKK884 pKa = 10.03IEE886 pKa = 3.99PTIFSHH892 pKa = 6.47MFPKK896 pKa = 10.43HH897 pKa = 6.13KK898 pKa = 10.7EE899 pKa = 3.76PVNLSKK905 pKa = 11.12YY906 pKa = 8.58GQHH909 pKa = 5.34TVKK912 pKa = 10.77DD913 pKa = 3.83VAKK916 pKa = 10.5KK917 pKa = 10.37SFSKK921 pKa = 10.73VKK923 pKa = 10.45EE924 pKa = 3.86ITQVEE929 pKa = 4.55LDD931 pKa = 3.74FGLDD935 pKa = 3.57YY936 pKa = 11.09LSTLMQPFSSLSEE949 pKa = 3.79QDD951 pKa = 3.34TVKK954 pKa = 10.95GFDD957 pKa = 3.24KK958 pKa = 10.67VAKK961 pKa = 10.22INMDD965 pKa = 2.85KK966 pKa = 10.08STGIGCEE973 pKa = 3.85RR974 pKa = 11.84DD975 pKa = 2.88RR976 pKa = 11.84AAYY979 pKa = 9.61INFEE983 pKa = 4.08RR984 pKa = 11.84GYY986 pKa = 8.85YY987 pKa = 8.05TQLLRR992 pKa = 11.84KK993 pKa = 8.78EE994 pKa = 4.23IKK996 pKa = 10.4DD997 pKa = 3.69IEE999 pKa = 4.76DD1000 pKa = 3.44NLLKK1004 pKa = 10.77DD1005 pKa = 3.29EE1006 pKa = 4.65HH1007 pKa = 7.45VNLNLWVAKK1016 pKa = 7.78EE1017 pKa = 3.97TLKK1020 pKa = 11.36DD1021 pKa = 3.6EE1022 pKa = 4.3VRR1024 pKa = 11.84SVSKK1028 pKa = 10.64DD1029 pKa = 3.13QEE1031 pKa = 4.03PRR1033 pKa = 11.84SFRR1036 pKa = 11.84VLRR1039 pKa = 11.84FPINVLCKK1047 pKa = 9.8QLTGEE1052 pKa = 4.2MVNNIIANKK1061 pKa = 8.93FSNGIMIGINPYY1073 pKa = 9.43SEE1075 pKa = 4.0WEE1077 pKa = 4.01TLYY1080 pKa = 9.89NTVGYY1085 pKa = 8.86KK1086 pKa = 10.27AKK1088 pKa = 10.75GIIAADD1094 pKa = 3.18IKK1096 pKa = 11.17KK1097 pKa = 10.12FDD1099 pKa = 4.01GGMLPQVQHH1108 pKa = 5.93GVVDD1112 pKa = 4.81TILSFFSGTYY1122 pKa = 9.98KK1123 pKa = 9.71EE1124 pKa = 4.46TEE1126 pKa = 3.95LLKK1129 pKa = 11.15GILHH1133 pKa = 6.14NFINNPVAVNDD1144 pKa = 3.89DD1145 pKa = 4.05VYY1147 pKa = 10.82LTTHH1151 pKa = 6.38SMPSGCYY1158 pKa = 8.15LTAIMNSMVQKK1169 pKa = 10.57VYY1171 pKa = 9.16TAMWYY1176 pKa = 10.14KK1177 pKa = 10.51HH1178 pKa = 6.37VIPDD1182 pKa = 3.64ATVTQYY1188 pKa = 11.34KK1189 pKa = 9.73QDD1191 pKa = 3.27VYY1193 pKa = 11.42DD1194 pKa = 4.46AVYY1197 pKa = 10.93GDD1199 pKa = 5.78DD1200 pKa = 4.19KK1201 pKa = 11.53LCSIKK1206 pKa = 10.89NKK1208 pKa = 10.16EE1209 pKa = 4.63DD1210 pKa = 3.13ILNAITMRR1218 pKa = 11.84DD1219 pKa = 3.59YY1220 pKa = 11.13FQLIGLDD1227 pKa = 4.07LSTADD1232 pKa = 4.35KK1233 pKa = 11.38KK1234 pKa = 11.23EE1235 pKa = 3.54ITQPFDD1241 pKa = 2.5SWEE1244 pKa = 3.99DD1245 pKa = 3.43VNFLKK1250 pKa = 10.61RR1251 pKa = 11.84KK1252 pKa = 9.0FVFHH1256 pKa = 6.8PVLKK1260 pKa = 10.35RR1261 pKa = 11.84IMCPLIDD1268 pKa = 3.51EE1269 pKa = 4.97TFYY1272 pKa = 12.01SMLAWWDD1279 pKa = 3.44TSKK1282 pKa = 11.37DD1283 pKa = 3.76FNDD1286 pKa = 4.15VIDD1289 pKa = 4.22GKK1291 pKa = 10.8LRR1293 pKa = 11.84SFTYY1297 pKa = 10.22EE1298 pKa = 3.51SYY1300 pKa = 10.98LRR1302 pKa = 11.84EE1303 pKa = 4.53DD1304 pKa = 3.31NDD1306 pKa = 4.32SNRR1309 pKa = 11.84EE1310 pKa = 3.88KK1311 pKa = 11.12LKK1313 pKa = 10.39MYY1315 pKa = 10.34LEE1317 pKa = 4.71NVAGMTWIMPSNMEE1331 pKa = 4.44MYY1333 pKa = 10.49RR1334 pKa = 11.84QFLKK1338 pKa = 11.3NEE1340 pKa = 3.71VDD1342 pKa = 3.57YY1343 pKa = 8.4TTYY1346 pKa = 11.43AA1347 pKa = 3.21
Molecular weight: 153.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2174 |
827 |
1347 |
1087.0 |
121.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.474 ± 0.535 | 1.472 ± 0.289 |
6.256 ± 0.328 | 3.864 ± 1.035 |
5.06 ± 0.335 | 5.52 ± 0.301 |
2.254 ± 0.233 | 6.624 ± 0.085 |
6.21 ± 2.032 | 9.108 ± 0.576 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.174 ± 0.017 | 5.244 ± 0.252 |
4.416 ± 1.279 | 3.542 ± 0.187 |
3.496 ± 0.145 | 8.832 ± 0.62 |
6.716 ± 0.932 | 7.82 ± 0.3 |
1.012 ± 0.233 | 3.91 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
