
Rhizobium sp. BK251
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
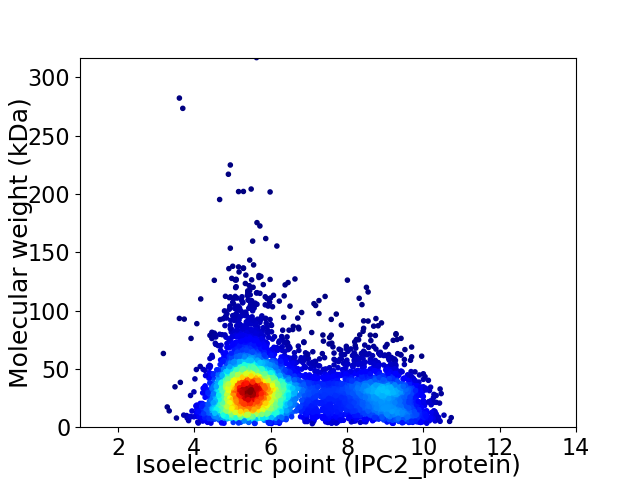
Virtual 2D-PAGE plot for 6346 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R1RUM2|A0A4R1RUM2_9RHIZ DNA-binding transcriptional LysR family regulator OS=Rhizobium sp. BK251 OX=2512125 GN=EV286_108267 PE=3 SV=1
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAVVSGAQAADD24 pKa = 4.21AIVAAEE30 pKa = 4.32PEE32 pKa = 4.04PVEE35 pKa = 4.04YY36 pKa = 11.08VRR38 pKa = 11.84VCDD41 pKa = 4.69AYY43 pKa = 9.35GTGYY47 pKa = 10.5FYY49 pKa = 10.55IPGTEE54 pKa = 3.7TCLQIGGWVRR64 pKa = 11.84FQVNYY69 pKa = 10.44GRR71 pKa = 11.84DD72 pKa = 3.25IAEE75 pKa = 4.28GSDD78 pKa = 3.45ANSSGRR84 pKa = 11.84GYY86 pKa = 10.87FYY88 pKa = 10.98LQTKK92 pKa = 10.32NDD94 pKa = 3.79TEE96 pKa = 4.44YY97 pKa = 9.76GTLTGFIAVEE107 pKa = 4.29GDD109 pKa = 3.48VSGQDD114 pKa = 3.29ASVNPYY120 pKa = 9.82FDD122 pKa = 3.28EE123 pKa = 6.48AYY125 pKa = 10.38LQLGGFKK132 pKa = 10.65AGWFYY137 pKa = 11.62NWWDD141 pKa = 3.19KK142 pKa = 11.36GINGEE147 pKa = 4.42QDD149 pKa = 3.37TLNSGGNEE157 pKa = 3.94WVSVSYY163 pKa = 9.79TYY165 pKa = 9.66EE166 pKa = 3.7TDD168 pKa = 3.05NFSIGGAVDD177 pKa = 3.33EE178 pKa = 5.0LDD180 pKa = 3.7RR181 pKa = 11.84DD182 pKa = 3.95GLSTVFAPGDD192 pKa = 4.23PINDD196 pKa = 3.72SNVVVEE202 pKa = 5.34DD203 pKa = 3.65SFDD206 pKa = 4.42GIDD209 pKa = 3.35YY210 pKa = 10.73GFEE213 pKa = 4.07GLITATFGGVSFDD226 pKa = 4.98LLGSYY231 pKa = 11.35DD232 pKa = 3.6NFNEE236 pKa = 3.88EE237 pKa = 3.85GAIRR241 pKa = 11.84ALMSAEE247 pKa = 4.52LGPGTLQAALMWASGQNVYY266 pKa = 10.43FDD268 pKa = 3.63EE269 pKa = 5.05SEE271 pKa = 3.83WSLAASYY278 pKa = 10.42AYY280 pKa = 9.5KK281 pKa = 11.02VNDD284 pKa = 4.36KK285 pKa = 9.84LTITPGGQYY294 pKa = 10.13FWQSSLDD301 pKa = 3.67DD302 pKa = 4.83DD303 pKa = 5.19GSFEE307 pKa = 6.38GGDD310 pKa = 3.16TWDD313 pKa = 3.77AGVTVDD319 pKa = 4.18YY320 pKa = 10.74RR321 pKa = 11.84ITEE324 pKa = 4.21GLTSKK329 pKa = 10.65ISVQYY334 pKa = 11.15NDD336 pKa = 5.45LGDD339 pKa = 4.14SDD341 pKa = 5.46DD342 pKa = 5.16DD343 pKa = 4.16SVSGFFRR350 pKa = 11.84LQRR353 pKa = 11.84TFF355 pKa = 3.23
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAVVSGAQAADD24 pKa = 4.21AIVAAEE30 pKa = 4.32PEE32 pKa = 4.04PVEE35 pKa = 4.04YY36 pKa = 11.08VRR38 pKa = 11.84VCDD41 pKa = 4.69AYY43 pKa = 9.35GTGYY47 pKa = 10.5FYY49 pKa = 10.55IPGTEE54 pKa = 3.7TCLQIGGWVRR64 pKa = 11.84FQVNYY69 pKa = 10.44GRR71 pKa = 11.84DD72 pKa = 3.25IAEE75 pKa = 4.28GSDD78 pKa = 3.45ANSSGRR84 pKa = 11.84GYY86 pKa = 10.87FYY88 pKa = 10.98LQTKK92 pKa = 10.32NDD94 pKa = 3.79TEE96 pKa = 4.44YY97 pKa = 9.76GTLTGFIAVEE107 pKa = 4.29GDD109 pKa = 3.48VSGQDD114 pKa = 3.29ASVNPYY120 pKa = 9.82FDD122 pKa = 3.28EE123 pKa = 6.48AYY125 pKa = 10.38LQLGGFKK132 pKa = 10.65AGWFYY137 pKa = 11.62NWWDD141 pKa = 3.19KK142 pKa = 11.36GINGEE147 pKa = 4.42QDD149 pKa = 3.37TLNSGGNEE157 pKa = 3.94WVSVSYY163 pKa = 9.79TYY165 pKa = 9.66EE166 pKa = 3.7TDD168 pKa = 3.05NFSIGGAVDD177 pKa = 3.33EE178 pKa = 5.0LDD180 pKa = 3.7RR181 pKa = 11.84DD182 pKa = 3.95GLSTVFAPGDD192 pKa = 4.23PINDD196 pKa = 3.72SNVVVEE202 pKa = 5.34DD203 pKa = 3.65SFDD206 pKa = 4.42GIDD209 pKa = 3.35YY210 pKa = 10.73GFEE213 pKa = 4.07GLITATFGGVSFDD226 pKa = 4.98LLGSYY231 pKa = 11.35DD232 pKa = 3.6NFNEE236 pKa = 3.88EE237 pKa = 3.85GAIRR241 pKa = 11.84ALMSAEE247 pKa = 4.52LGPGTLQAALMWASGQNVYY266 pKa = 10.43FDD268 pKa = 3.63EE269 pKa = 5.05SEE271 pKa = 3.83WSLAASYY278 pKa = 10.42AYY280 pKa = 9.5KK281 pKa = 11.02VNDD284 pKa = 4.36KK285 pKa = 9.84LTITPGGQYY294 pKa = 10.13FWQSSLDD301 pKa = 3.67DD302 pKa = 4.83DD303 pKa = 5.19GSFEE307 pKa = 6.38GGDD310 pKa = 3.16TWDD313 pKa = 3.77AGVTVDD319 pKa = 4.18YY320 pKa = 10.74RR321 pKa = 11.84ITEE324 pKa = 4.21GLTSKK329 pKa = 10.65ISVQYY334 pKa = 11.15NDD336 pKa = 5.45LGDD339 pKa = 4.14SDD341 pKa = 5.46DD342 pKa = 5.16DD343 pKa = 4.16SVSGFFRR350 pKa = 11.84LQRR353 pKa = 11.84TFF355 pKa = 3.23
Molecular weight: 38.41 kDa
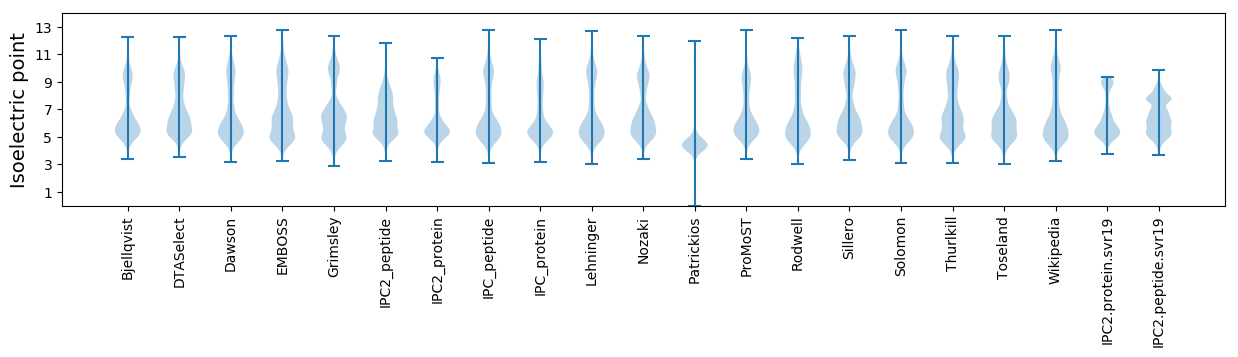
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R1RGA3|A0A4R1RGA3_9RHIZ Response regulator receiver domain-containing protein OS=Rhizobium sp. BK251 OX=2512125 GN=EV286_11453 PE=4 SV=1
MM1 pKa = 7.19SRR3 pKa = 11.84IHH5 pKa = 6.66AATRR9 pKa = 11.84LKK11 pKa = 10.02STLRR15 pKa = 11.84RR16 pKa = 11.84CLPSGGPCVGRR27 pKa = 11.84CRR29 pKa = 11.84GIRR32 pKa = 11.84DD33 pKa = 3.29ILVFPVMSAAEE44 pKa = 4.49GEE46 pKa = 4.46VSSPIMAGMSFRR58 pKa = 11.84MRR60 pKa = 11.84LHH62 pKa = 6.55GEE64 pKa = 3.66NKK66 pKa = 9.67GRR68 pKa = 11.84STFIRR73 pKa = 11.84SASPRR78 pKa = 11.84RR79 pKa = 11.84SFGRR83 pKa = 11.84KK84 pKa = 6.98QRR86 pKa = 11.84TGGIAFLSIFGRR98 pKa = 11.84ATLNLPDD105 pKa = 4.79RR106 pKa = 11.84GCGPLRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84DD115 pKa = 3.64GEE117 pKa = 4.47PIFAEE122 pKa = 4.41PWHH125 pKa = 5.76AQIMAIADD133 pKa = 4.04LLIGSGPIPQSQWTQTLGKK152 pKa = 8.75EE153 pKa = 3.85LRR155 pKa = 11.84LARR158 pKa = 11.84LADD161 pKa = 3.84SADD164 pKa = 3.72DD165 pKa = 3.34TSTYY169 pKa = 8.29YY170 pKa = 10.42TAVLSALEE178 pKa = 4.72RR179 pKa = 11.84ILDD182 pKa = 3.7AGGNVSCVEE191 pKa = 3.76LARR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84EE197 pKa = 4.1EE198 pKa = 3.38WRR200 pKa = 11.84QAYY203 pKa = 9.72LHH205 pKa = 5.7THH207 pKa = 6.52HH208 pKa = 6.62GQPVEE213 pKa = 4.08LEE215 pKa = 3.92SRR217 pKa = 11.84FAKK220 pKa = 10.24TSDD223 pKa = 2.94
MM1 pKa = 7.19SRR3 pKa = 11.84IHH5 pKa = 6.66AATRR9 pKa = 11.84LKK11 pKa = 10.02STLRR15 pKa = 11.84RR16 pKa = 11.84CLPSGGPCVGRR27 pKa = 11.84CRR29 pKa = 11.84GIRR32 pKa = 11.84DD33 pKa = 3.29ILVFPVMSAAEE44 pKa = 4.49GEE46 pKa = 4.46VSSPIMAGMSFRR58 pKa = 11.84MRR60 pKa = 11.84LHH62 pKa = 6.55GEE64 pKa = 3.66NKK66 pKa = 9.67GRR68 pKa = 11.84STFIRR73 pKa = 11.84SASPRR78 pKa = 11.84RR79 pKa = 11.84SFGRR83 pKa = 11.84KK84 pKa = 6.98QRR86 pKa = 11.84TGGIAFLSIFGRR98 pKa = 11.84ATLNLPDD105 pKa = 4.79RR106 pKa = 11.84GCGPLRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84DD115 pKa = 3.64GEE117 pKa = 4.47PIFAEE122 pKa = 4.41PWHH125 pKa = 5.76AQIMAIADD133 pKa = 4.04LLIGSGPIPQSQWTQTLGKK152 pKa = 8.75EE153 pKa = 3.85LRR155 pKa = 11.84LARR158 pKa = 11.84LADD161 pKa = 3.84SADD164 pKa = 3.72DD165 pKa = 3.34TSTYY169 pKa = 8.29YY170 pKa = 10.42TAVLSALEE178 pKa = 4.72RR179 pKa = 11.84ILDD182 pKa = 3.7AGGNVSCVEE191 pKa = 3.76LARR194 pKa = 11.84RR195 pKa = 11.84RR196 pKa = 11.84EE197 pKa = 4.1EE198 pKa = 3.38WRR200 pKa = 11.84QAYY203 pKa = 9.72LHH205 pKa = 5.7THH207 pKa = 6.52HH208 pKa = 6.62GQPVEE213 pKa = 4.08LEE215 pKa = 3.92SRR217 pKa = 11.84FAKK220 pKa = 10.24TSDD223 pKa = 2.94
Molecular weight: 24.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1948081 |
29 |
2834 |
307.0 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.847 ± 0.04 | 0.79 ± 0.008 |
5.654 ± 0.027 | 5.844 ± 0.028 |
3.925 ± 0.018 | 8.348 ± 0.033 |
2.039 ± 0.014 | 5.653 ± 0.024 |
3.54 ± 0.025 | 10.002 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.53 ± 0.014 | 2.847 ± 0.018 |
4.929 ± 0.022 | 3.084 ± 0.018 |
6.806 ± 0.027 | 5.863 ± 0.023 |
5.269 ± 0.02 | 7.391 ± 0.026 |
1.31 ± 0.012 | 2.328 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
