
Gibberella fujikuroi (strain CBS 195.34 / IMI 58289 / NRRL A-6831) (Bakanae and foot rot disease fungus) (Fusarium fujikuroi)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Nectriaceae; Fusarium; Fusarium fujikuroi species complex; Fusarium fujikuroi
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
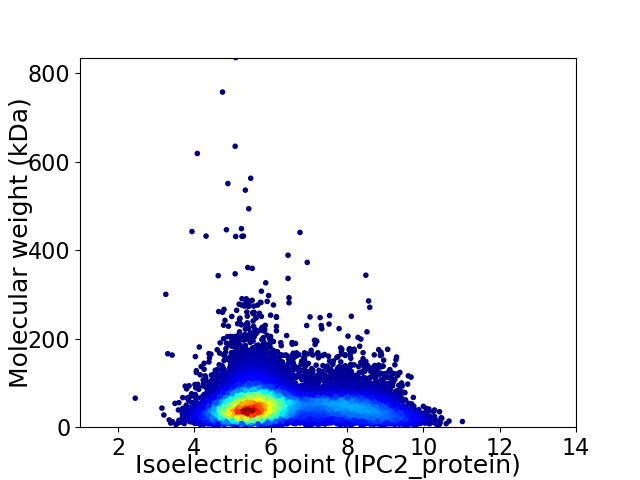
Virtual 2D-PAGE plot for 14792 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S0DWK4|S0DWK4_GIBF5 Uncharacterized protein OS=Gibberella fujikuroi (strain CBS 195.34 / IMI 58289 / NRRL A-6831) OX=1279085 GN=FFUJ_13049 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.43SVLTFAGLAFASFAAAGPCRR22 pKa = 11.84PRR24 pKa = 11.84TTTTAAAVSSSTEE37 pKa = 3.94IASSTATEE45 pKa = 4.03TAAVSTSTGYY55 pKa = 10.21SVPVVSEE62 pKa = 4.14SSTTLATVIVSEE74 pKa = 4.53TSATEE79 pKa = 3.85TSAGEE84 pKa = 4.03TSATGTTTAEE94 pKa = 3.87TSAAGTTTADD104 pKa = 3.26TSAVDD109 pKa = 3.8TTTTAADD116 pKa = 3.5TTTTVEE122 pKa = 4.24GTTTTVADD130 pKa = 3.79TTTAAEE136 pKa = 4.32DD137 pKa = 3.96TTTTAADD144 pKa = 3.65TTTTAEE150 pKa = 4.14GTTTAEE156 pKa = 4.16GTTTEE161 pKa = 4.3GTTTAAEE168 pKa = 4.3TTTTAEE174 pKa = 4.3GTTTTAEE181 pKa = 4.13DD182 pKa = 3.65TTTAEE187 pKa = 4.15TTTAEE192 pKa = 4.42GTTTAADD199 pKa = 3.66TTTTAEE205 pKa = 4.33GTTTTAEE212 pKa = 4.18GTTTADD218 pKa = 3.2ATTSTSAEE226 pKa = 3.9PTADD230 pKa = 3.44QSCDD234 pKa = 3.45NAGLEE239 pKa = 4.1YY240 pKa = 10.43AIYY243 pKa = 10.63KK244 pKa = 7.96HH245 pKa = 5.67TFYY248 pKa = 11.24NSDD251 pKa = 3.58PPHH254 pKa = 6.65FSSFDD259 pKa = 3.19PTFFHH264 pKa = 6.67TATPTYY270 pKa = 9.93QGEE273 pKa = 4.39TTRR276 pKa = 11.84IGIPPGTASDD286 pKa = 3.67TSFAIYY292 pKa = 10.25DD293 pKa = 3.97GSPSQLWQYY302 pKa = 10.82KK303 pKa = 9.4AVNHH307 pKa = 6.28RR308 pKa = 11.84AFLYY312 pKa = 10.7APEE315 pKa = 4.48TGDD318 pKa = 3.43YY319 pKa = 10.1TVTIPNSDD327 pKa = 4.63EE328 pKa = 3.6ITLIWFGEE336 pKa = 4.16KK337 pKa = 10.04AIEE340 pKa = 4.12GWTRR344 pKa = 11.84ANADD348 pKa = 4.03LEE350 pKa = 4.0QDD352 pKa = 4.0YY353 pKa = 10.87PGGTSKK359 pKa = 9.61TFTIHH364 pKa = 5.93LTAGTYY370 pKa = 9.46TPFRR374 pKa = 11.84LLWANAQGDD383 pKa = 4.42LNFIAEE389 pKa = 4.19VQAPDD394 pKa = 3.22GKK396 pKa = 11.2VIVNGDD402 pKa = 3.32GSDD405 pKa = 2.8NKK407 pKa = 10.48YY408 pKa = 10.16FVRR411 pKa = 11.84FACDD415 pKa = 2.79EE416 pKa = 4.32STPSFPDD423 pKa = 3.52FGYY426 pKa = 11.13GGG428 pKa = 3.49
MM1 pKa = 7.62KK2 pKa = 10.43SVLTFAGLAFASFAAAGPCRR22 pKa = 11.84PRR24 pKa = 11.84TTTTAAAVSSSTEE37 pKa = 3.94IASSTATEE45 pKa = 4.03TAAVSTSTGYY55 pKa = 10.21SVPVVSEE62 pKa = 4.14SSTTLATVIVSEE74 pKa = 4.53TSATEE79 pKa = 3.85TSAGEE84 pKa = 4.03TSATGTTTAEE94 pKa = 3.87TSAAGTTTADD104 pKa = 3.26TSAVDD109 pKa = 3.8TTTTAADD116 pKa = 3.5TTTTVEE122 pKa = 4.24GTTTTVADD130 pKa = 3.79TTTAAEE136 pKa = 4.32DD137 pKa = 3.96TTTTAADD144 pKa = 3.65TTTTAEE150 pKa = 4.14GTTTAEE156 pKa = 4.16GTTTEE161 pKa = 4.3GTTTAAEE168 pKa = 4.3TTTTAEE174 pKa = 4.3GTTTTAEE181 pKa = 4.13DD182 pKa = 3.65TTTAEE187 pKa = 4.15TTTAEE192 pKa = 4.42GTTTAADD199 pKa = 3.66TTTTAEE205 pKa = 4.33GTTTTAEE212 pKa = 4.18GTTTADD218 pKa = 3.2ATTSTSAEE226 pKa = 3.9PTADD230 pKa = 3.44QSCDD234 pKa = 3.45NAGLEE239 pKa = 4.1YY240 pKa = 10.43AIYY243 pKa = 10.63KK244 pKa = 7.96HH245 pKa = 5.67TFYY248 pKa = 11.24NSDD251 pKa = 3.58PPHH254 pKa = 6.65FSSFDD259 pKa = 3.19PTFFHH264 pKa = 6.67TATPTYY270 pKa = 9.93QGEE273 pKa = 4.39TTRR276 pKa = 11.84IGIPPGTASDD286 pKa = 3.67TSFAIYY292 pKa = 10.25DD293 pKa = 3.97GSPSQLWQYY302 pKa = 10.82KK303 pKa = 9.4AVNHH307 pKa = 6.28RR308 pKa = 11.84AFLYY312 pKa = 10.7APEE315 pKa = 4.48TGDD318 pKa = 3.43YY319 pKa = 10.1TVTIPNSDD327 pKa = 4.63EE328 pKa = 3.6ITLIWFGEE336 pKa = 4.16KK337 pKa = 10.04AIEE340 pKa = 4.12GWTRR344 pKa = 11.84ANADD348 pKa = 4.03LEE350 pKa = 4.0QDD352 pKa = 4.0YY353 pKa = 10.87PGGTSKK359 pKa = 9.61TFTIHH364 pKa = 5.93LTAGTYY370 pKa = 9.46TPFRR374 pKa = 11.84LLWANAQGDD383 pKa = 4.42LNFIAEE389 pKa = 4.19VQAPDD394 pKa = 3.22GKK396 pKa = 11.2VIVNGDD402 pKa = 3.32GSDD405 pKa = 2.8NKK407 pKa = 10.48YY408 pKa = 10.16FVRR411 pKa = 11.84FACDD415 pKa = 2.79EE416 pKa = 4.32STPSFPDD423 pKa = 3.52FGYY426 pKa = 11.13GGG428 pKa = 3.49
Molecular weight: 43.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S0E214|S0E214_GIBF5 Related to transcriptional coactivator HFI1 OS=Gibberella fujikuroi (strain CBS 195.34 / IMI 58289 / NRRL A-6831) OX=1279085 GN=FFUJ_07552 PE=4 SV=1
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APVHH26 pKa = 4.79SHH28 pKa = 5.43RR29 pKa = 11.84HH30 pKa = 4.01TTTTTTTTTKK40 pKa = 9.5PRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GMFGGGASRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 5.59ATATAPVHH64 pKa = 4.93HH65 pKa = 6.66HH66 pKa = 5.18QRR68 pKa = 11.84RR69 pKa = 11.84PSMKK73 pKa = 10.02DD74 pKa = 2.95KK75 pKa = 11.41VSGALLKK82 pKa = 11.04LKK84 pKa = 10.68GSLTRR89 pKa = 11.84RR90 pKa = 11.84PGVKK94 pKa = 9.89AAGTRR99 pKa = 11.84RR100 pKa = 11.84MRR102 pKa = 11.84GTDD105 pKa = 3.02GRR107 pKa = 11.84GARR110 pKa = 11.84HH111 pKa = 5.87HH112 pKa = 7.23RR113 pKa = 11.84YY114 pKa = 9.44
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APVHH26 pKa = 4.79SHH28 pKa = 5.43RR29 pKa = 11.84HH30 pKa = 4.01TTTTTTTTTKK40 pKa = 9.5PRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GMFGGGASRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 5.59ATATAPVHH64 pKa = 4.93HH65 pKa = 6.66HH66 pKa = 5.18QRR68 pKa = 11.84RR69 pKa = 11.84PSMKK73 pKa = 10.02DD74 pKa = 2.95KK75 pKa = 11.41VSGALLKK82 pKa = 11.04LKK84 pKa = 10.68GSLTRR89 pKa = 11.84RR90 pKa = 11.84PGVKK94 pKa = 9.89AAGTRR99 pKa = 11.84RR100 pKa = 11.84MRR102 pKa = 11.84GTDD105 pKa = 3.02GRR107 pKa = 11.84GARR110 pKa = 11.84HH111 pKa = 5.87HH112 pKa = 7.23RR113 pKa = 11.84YY114 pKa = 9.44
Molecular weight: 12.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7170111 |
23 |
7581 |
484.7 |
53.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.3 ± 0.019 | 1.331 ± 0.008 |
5.862 ± 0.016 | 6.207 ± 0.019 |
3.803 ± 0.014 | 6.748 ± 0.019 |
2.4 ± 0.008 | 5.16 ± 0.013 |
5.039 ± 0.019 | 8.866 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.224 ± 0.008 | 3.776 ± 0.009 |
5.86 ± 0.023 | 4.004 ± 0.014 |
5.829 ± 0.016 | 8.121 ± 0.025 |
6.009 ± 0.026 | 6.092 ± 0.013 |
1.552 ± 0.007 | 2.818 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
