
Aureitalea marina
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aureitalea
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
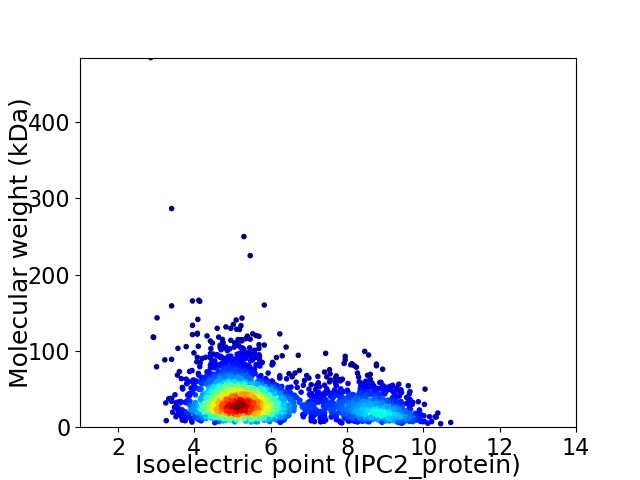
Virtual 2D-PAGE plot for 2645 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S7KSY9|A0A2S7KSY9_9FLAO Uncharacterized protein OS=Aureitalea marina OX=930804 GN=BST85_12915 PE=4 SV=1
MM1 pKa = 7.71KK2 pKa = 10.25LQLSLLFVLALISSGVSAQQVSSEE26 pKa = 4.05GAFYY30 pKa = 9.74TGQVASMRR38 pKa = 11.84YY39 pKa = 9.22VEE41 pKa = 4.5PLSSRR46 pKa = 11.84TNLIPAEE53 pKa = 4.27DD54 pKa = 3.83RR55 pKa = 11.84TGEE58 pKa = 4.08AQDD61 pKa = 3.91GRR63 pKa = 11.84SSRR66 pKa = 11.84PEE68 pKa = 3.97VIPGKK73 pKa = 10.29DD74 pKa = 3.16PQTTNDD80 pKa = 3.52YY81 pKa = 9.97FASNPHH87 pKa = 6.56PMTGKK92 pKa = 9.54IASRR96 pKa = 11.84SPLIVFDD103 pKa = 5.03ASASSSQPTDD113 pKa = 3.16PSLAVGPNHH122 pKa = 6.66VISVFNTGFVIFNDD136 pKa = 3.86KK137 pKa = 10.75LDD139 pKa = 4.21PANNNTGQLSVTNIFSGGGCCDD161 pKa = 3.2LTVSYY166 pKa = 11.07DD167 pKa = 3.33SAADD171 pKa = 3.63RR172 pKa = 11.84WVLSYY177 pKa = 11.26LFVGAGAEE185 pKa = 4.18IAVSDD190 pKa = 4.53GPDD193 pKa = 3.41PNTAGWYY200 pKa = 9.62VYY202 pKa = 10.16SIPQINDD209 pKa = 3.22YY210 pKa = 11.02QKK212 pKa = 11.18LSVWSDD218 pKa = 2.88GYY220 pKa = 11.46YY221 pKa = 9.04ITDD224 pKa = 3.25NVGGSNRR231 pKa = 11.84VWALEE236 pKa = 3.66RR237 pKa = 11.84DD238 pKa = 3.62ALLAGDD244 pKa = 4.62PAASIQGFNLPGIVTSGFFSPQALNVTDD272 pKa = 5.46DD273 pKa = 3.87NMPADD278 pKa = 4.09GNATIIYY285 pKa = 8.97LQDD288 pKa = 4.04DD289 pKa = 3.62AWSGVSFDD297 pKa = 4.68HH298 pKa = 6.67FKK300 pKa = 10.33IWTIDD305 pKa = 3.26ADD307 pKa = 3.96FDD309 pKa = 4.06NPGNSTVSAPTEE321 pKa = 3.73IATDD325 pKa = 3.63PFISVFDD332 pKa = 3.97GGSFSNLTQPGGGVAIDD349 pKa = 3.99ALQATIMNQAQFRR362 pKa = 11.84KK363 pKa = 9.39FPAYY367 pKa = 10.51NSALFNFVVDD377 pKa = 3.52VDD379 pKa = 3.76ATSGEE384 pKa = 4.24KK385 pKa = 10.53AGVRR389 pKa = 11.84WFEE392 pKa = 3.86LRR394 pKa = 11.84QDD396 pKa = 3.65GDD398 pKa = 3.87GQPWTVYY405 pKa = 10.48QEE407 pKa = 4.27GTYY410 pKa = 9.69TSPDD414 pKa = 3.45TPTGGEE420 pKa = 3.59KK421 pKa = 10.27HH422 pKa = 6.16AWHH425 pKa = 7.26ASLMMDD431 pKa = 3.63GSGNIGMGYY440 pKa = 10.03SAMTVDD446 pKa = 4.8PSDD449 pKa = 4.12PNPIRR454 pKa = 11.84VSSYY458 pKa = 7.5YY459 pKa = 8.61TGRR462 pKa = 11.84YY463 pKa = 9.47ASDD466 pKa = 3.59PLGTMTIEE474 pKa = 4.26EE475 pKa = 4.09EE476 pKa = 4.79LIANGNANIPGLRR489 pKa = 11.84YY490 pKa = 10.05GDD492 pKa = 3.79YY493 pKa = 11.03SKK495 pKa = 11.02IDD497 pKa = 3.45IDD499 pKa = 3.91PVNDD503 pKa = 3.2KK504 pKa = 9.31TFWFINEE511 pKa = 3.9YY512 pKa = 9.77MNSGRR517 pKa = 11.84KK518 pKa = 8.85NVVGVFQITPNFNNDD533 pKa = 3.21AGVVSIDD540 pKa = 3.64TPVTGTLSDD549 pKa = 3.85SEE551 pKa = 4.93EE552 pKa = 3.8ITVTIFNYY560 pKa = 10.59GLDD563 pKa = 4.36PIADD567 pKa = 3.7FDD569 pKa = 3.59VTYY572 pKa = 10.59QVDD575 pKa = 3.5GGAVVTEE582 pKa = 4.39TFNGTIEE589 pKa = 4.25STEE592 pKa = 3.87TAQFSFVTLADD603 pKa = 4.15LSTVGQTYY611 pKa = 10.66SITASTALAGDD622 pKa = 3.9EE623 pKa = 5.25DD624 pKa = 4.25NGNDD628 pKa = 3.45STTKK632 pKa = 10.1EE633 pKa = 4.15VTFLEE638 pKa = 4.66PNDD641 pKa = 4.36LGVTAITAPQSGTNLPADD659 pKa = 3.68AEE661 pKa = 4.33ITVTITNFGGEE672 pKa = 4.29SQSNFDD678 pKa = 2.99VTYY681 pKa = 10.69NLDD684 pKa = 3.61GNEE687 pKa = 3.84VTEE690 pKa = 4.39QVAGPLAGNSEE701 pKa = 4.27LSYY704 pKa = 10.94TFTQTADD711 pKa = 3.22LSAFGSYY718 pKa = 10.32SLSARR723 pKa = 11.84TDD725 pKa = 3.32LPTDD729 pKa = 3.36SDD731 pKa = 3.74NTNDD735 pKa = 3.36EE736 pKa = 4.31TAVTIVNSNCEE747 pKa = 3.85PGANCTLGDD756 pKa = 4.22GIVRR760 pKa = 11.84LQLEE764 pKa = 4.38NVDD767 pKa = 4.26NNSGCDD773 pKa = 3.33PDD775 pKa = 5.94GYY777 pKa = 11.65GNYY780 pKa = 9.4TNLIVEE786 pKa = 5.23LPTPDD791 pKa = 4.2ADD793 pKa = 3.85YY794 pKa = 11.48DD795 pKa = 3.9LTITTGYY802 pKa = 10.46GNQFVRR808 pKa = 11.84AWIDD812 pKa = 3.48FNDD815 pKa = 4.09DD816 pKa = 3.41FVFTIDD822 pKa = 3.49EE823 pKa = 4.34LVVNNYY829 pKa = 9.61EE830 pKa = 4.01IADD833 pKa = 3.82GQGAGDD839 pKa = 3.73YY840 pKa = 9.83TEE842 pKa = 4.68TMTLSVPASASPGQHH857 pKa = 6.23LMRR860 pKa = 11.84VKK862 pKa = 9.92TNWNAPVPDD871 pKa = 4.61DD872 pKa = 3.71ACEE875 pKa = 3.71EE876 pKa = 4.23TTYY879 pKa = 11.58GEE881 pKa = 4.34TEE883 pKa = 4.59DD884 pKa = 3.92YY885 pKa = 10.56LAEE888 pKa = 4.78IGVLSFGDD896 pKa = 3.56NQVSSEE902 pKa = 4.21DD903 pKa = 3.71LVIIHH908 pKa = 6.17QGNNLFEE915 pKa = 4.36VTMISPNGPEE925 pKa = 3.89KK926 pKa = 11.21LEE928 pKa = 3.94LSVYY932 pKa = 10.53NILGQQILNRR942 pKa = 11.84RR943 pKa = 11.84LEE945 pKa = 4.61SNNGTYY951 pKa = 10.46NYY953 pKa = 9.04QLNMNYY959 pKa = 9.61VSSGVYY965 pKa = 7.83MVRR968 pKa = 11.84LGTSQGGLVKK978 pKa = 10.53KK979 pKa = 10.54IVVRR983 pKa = 4.19
MM1 pKa = 7.71KK2 pKa = 10.25LQLSLLFVLALISSGVSAQQVSSEE26 pKa = 4.05GAFYY30 pKa = 9.74TGQVASMRR38 pKa = 11.84YY39 pKa = 9.22VEE41 pKa = 4.5PLSSRR46 pKa = 11.84TNLIPAEE53 pKa = 4.27DD54 pKa = 3.83RR55 pKa = 11.84TGEE58 pKa = 4.08AQDD61 pKa = 3.91GRR63 pKa = 11.84SSRR66 pKa = 11.84PEE68 pKa = 3.97VIPGKK73 pKa = 10.29DD74 pKa = 3.16PQTTNDD80 pKa = 3.52YY81 pKa = 9.97FASNPHH87 pKa = 6.56PMTGKK92 pKa = 9.54IASRR96 pKa = 11.84SPLIVFDD103 pKa = 5.03ASASSSQPTDD113 pKa = 3.16PSLAVGPNHH122 pKa = 6.66VISVFNTGFVIFNDD136 pKa = 3.86KK137 pKa = 10.75LDD139 pKa = 4.21PANNNTGQLSVTNIFSGGGCCDD161 pKa = 3.2LTVSYY166 pKa = 11.07DD167 pKa = 3.33SAADD171 pKa = 3.63RR172 pKa = 11.84WVLSYY177 pKa = 11.26LFVGAGAEE185 pKa = 4.18IAVSDD190 pKa = 4.53GPDD193 pKa = 3.41PNTAGWYY200 pKa = 9.62VYY202 pKa = 10.16SIPQINDD209 pKa = 3.22YY210 pKa = 11.02QKK212 pKa = 11.18LSVWSDD218 pKa = 2.88GYY220 pKa = 11.46YY221 pKa = 9.04ITDD224 pKa = 3.25NVGGSNRR231 pKa = 11.84VWALEE236 pKa = 3.66RR237 pKa = 11.84DD238 pKa = 3.62ALLAGDD244 pKa = 4.62PAASIQGFNLPGIVTSGFFSPQALNVTDD272 pKa = 5.46DD273 pKa = 3.87NMPADD278 pKa = 4.09GNATIIYY285 pKa = 8.97LQDD288 pKa = 4.04DD289 pKa = 3.62AWSGVSFDD297 pKa = 4.68HH298 pKa = 6.67FKK300 pKa = 10.33IWTIDD305 pKa = 3.26ADD307 pKa = 3.96FDD309 pKa = 4.06NPGNSTVSAPTEE321 pKa = 3.73IATDD325 pKa = 3.63PFISVFDD332 pKa = 3.97GGSFSNLTQPGGGVAIDD349 pKa = 3.99ALQATIMNQAQFRR362 pKa = 11.84KK363 pKa = 9.39FPAYY367 pKa = 10.51NSALFNFVVDD377 pKa = 3.52VDD379 pKa = 3.76ATSGEE384 pKa = 4.24KK385 pKa = 10.53AGVRR389 pKa = 11.84WFEE392 pKa = 3.86LRR394 pKa = 11.84QDD396 pKa = 3.65GDD398 pKa = 3.87GQPWTVYY405 pKa = 10.48QEE407 pKa = 4.27GTYY410 pKa = 9.69TSPDD414 pKa = 3.45TPTGGEE420 pKa = 3.59KK421 pKa = 10.27HH422 pKa = 6.16AWHH425 pKa = 7.26ASLMMDD431 pKa = 3.63GSGNIGMGYY440 pKa = 10.03SAMTVDD446 pKa = 4.8PSDD449 pKa = 4.12PNPIRR454 pKa = 11.84VSSYY458 pKa = 7.5YY459 pKa = 8.61TGRR462 pKa = 11.84YY463 pKa = 9.47ASDD466 pKa = 3.59PLGTMTIEE474 pKa = 4.26EE475 pKa = 4.09EE476 pKa = 4.79LIANGNANIPGLRR489 pKa = 11.84YY490 pKa = 10.05GDD492 pKa = 3.79YY493 pKa = 11.03SKK495 pKa = 11.02IDD497 pKa = 3.45IDD499 pKa = 3.91PVNDD503 pKa = 3.2KK504 pKa = 9.31TFWFINEE511 pKa = 3.9YY512 pKa = 9.77MNSGRR517 pKa = 11.84KK518 pKa = 8.85NVVGVFQITPNFNNDD533 pKa = 3.21AGVVSIDD540 pKa = 3.64TPVTGTLSDD549 pKa = 3.85SEE551 pKa = 4.93EE552 pKa = 3.8ITVTIFNYY560 pKa = 10.59GLDD563 pKa = 4.36PIADD567 pKa = 3.7FDD569 pKa = 3.59VTYY572 pKa = 10.59QVDD575 pKa = 3.5GGAVVTEE582 pKa = 4.39TFNGTIEE589 pKa = 4.25STEE592 pKa = 3.87TAQFSFVTLADD603 pKa = 4.15LSTVGQTYY611 pKa = 10.66SITASTALAGDD622 pKa = 3.9EE623 pKa = 5.25DD624 pKa = 4.25NGNDD628 pKa = 3.45STTKK632 pKa = 10.1EE633 pKa = 4.15VTFLEE638 pKa = 4.66PNDD641 pKa = 4.36LGVTAITAPQSGTNLPADD659 pKa = 3.68AEE661 pKa = 4.33ITVTITNFGGEE672 pKa = 4.29SQSNFDD678 pKa = 2.99VTYY681 pKa = 10.69NLDD684 pKa = 3.61GNEE687 pKa = 3.84VTEE690 pKa = 4.39QVAGPLAGNSEE701 pKa = 4.27LSYY704 pKa = 10.94TFTQTADD711 pKa = 3.22LSAFGSYY718 pKa = 10.32SLSARR723 pKa = 11.84TDD725 pKa = 3.32LPTDD729 pKa = 3.36SDD731 pKa = 3.74NTNDD735 pKa = 3.36EE736 pKa = 4.31TAVTIVNSNCEE747 pKa = 3.85PGANCTLGDD756 pKa = 4.22GIVRR760 pKa = 11.84LQLEE764 pKa = 4.38NVDD767 pKa = 4.26NNSGCDD773 pKa = 3.33PDD775 pKa = 5.94GYY777 pKa = 11.65GNYY780 pKa = 9.4TNLIVEE786 pKa = 5.23LPTPDD791 pKa = 4.2ADD793 pKa = 3.85YY794 pKa = 11.48DD795 pKa = 3.9LTITTGYY802 pKa = 10.46GNQFVRR808 pKa = 11.84AWIDD812 pKa = 3.48FNDD815 pKa = 4.09DD816 pKa = 3.41FVFTIDD822 pKa = 3.49EE823 pKa = 4.34LVVNNYY829 pKa = 9.61EE830 pKa = 4.01IADD833 pKa = 3.82GQGAGDD839 pKa = 3.73YY840 pKa = 9.83TEE842 pKa = 4.68TMTLSVPASASPGQHH857 pKa = 6.23LMRR860 pKa = 11.84VKK862 pKa = 9.92TNWNAPVPDD871 pKa = 4.61DD872 pKa = 3.71ACEE875 pKa = 3.71EE876 pKa = 4.23TTYY879 pKa = 11.58GEE881 pKa = 4.34TEE883 pKa = 4.59DD884 pKa = 3.92YY885 pKa = 10.56LAEE888 pKa = 4.78IGVLSFGDD896 pKa = 3.56NQVSSEE902 pKa = 4.21DD903 pKa = 3.71LVIIHH908 pKa = 6.17QGNNLFEE915 pKa = 4.36VTMISPNGPEE925 pKa = 3.89KK926 pKa = 11.21LEE928 pKa = 3.94LSVYY932 pKa = 10.53NILGQQILNRR942 pKa = 11.84RR943 pKa = 11.84LEE945 pKa = 4.61SNNGTYY951 pKa = 10.46NYY953 pKa = 9.04QLNMNYY959 pKa = 9.61VSSGVYY965 pKa = 7.83MVRR968 pKa = 11.84LGTSQGGLVKK978 pKa = 10.53KK979 pKa = 10.54IVVRR983 pKa = 4.19
Molecular weight: 105.59 kDa
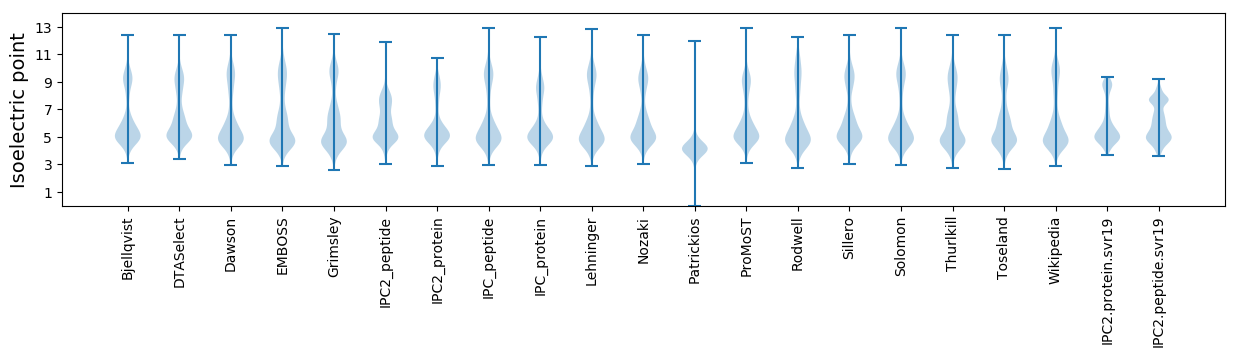
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S7KPT9|A0A2S7KPT9_9FLAO Uncharacterized protein OS=Aureitalea marina OX=930804 GN=BST85_06865 PE=3 SV=1
MM1 pKa = 7.66ISDD4 pKa = 4.0GTNRR8 pKa = 11.84RR9 pKa = 11.84STVLLILGLLLCFLANISAGSVHH32 pKa = 6.49VPLEE36 pKa = 4.31DD37 pKa = 2.96VWQGLIGGEE46 pKa = 4.23TSRR49 pKa = 11.84PAYY52 pKa = 10.16RR53 pKa = 11.84FIILEE58 pKa = 3.78YY59 pKa = 9.87RR60 pKa = 11.84LPKK63 pKa = 10.46AITAVLTGSGLAVSGLLMQTLFRR86 pKa = 11.84NPLAGPFVLGLSSGASLGVAILILGAGTFGGWLGGLLLSNWSLVLASALGSFLVLMAVLAVTLRR150 pKa = 11.84VKK152 pKa = 10.24DD153 pKa = 3.57TMAILIIGLMFGSVTASVVGVLSYY177 pKa = 9.09FSPAEE182 pKa = 3.67QLQQYY187 pKa = 7.02VFWSFGSLGNLSWQALGVLTLCWLFGMILAIGRR220 pKa = 11.84IKK222 pKa = 10.73SLNSLLLGTRR232 pKa = 11.84YY233 pKa = 9.38AQSMGLRR240 pKa = 11.84VRR242 pKa = 11.84RR243 pKa = 11.84TTLLLIVATSLLAGGITAFAGPIAFVGLAVPHH275 pKa = 6.46LTRR278 pKa = 11.84QLVPSADD285 pKa = 3.35HH286 pKa = 6.94RR287 pKa = 11.84LLLPAVVLFGAALMLICDD305 pKa = 5.37TIAQLPGSEE314 pKa = 3.99FTLPINAITSLVGAPVVIWLLVRR337 pKa = 11.84KK338 pKa = 9.59RR339 pKa = 11.84KK340 pKa = 10.16LMFF343 pKa = 4.27
MM1 pKa = 7.66ISDD4 pKa = 4.0GTNRR8 pKa = 11.84RR9 pKa = 11.84STVLLILGLLLCFLANISAGSVHH32 pKa = 6.49VPLEE36 pKa = 4.31DD37 pKa = 2.96VWQGLIGGEE46 pKa = 4.23TSRR49 pKa = 11.84PAYY52 pKa = 10.16RR53 pKa = 11.84FIILEE58 pKa = 3.78YY59 pKa = 9.87RR60 pKa = 11.84LPKK63 pKa = 10.46AITAVLTGSGLAVSGLLMQTLFRR86 pKa = 11.84NPLAGPFVLGLSSGASLGVAILILGAGTFGGWLGGLLLSNWSLVLASALGSFLVLMAVLAVTLRR150 pKa = 11.84VKK152 pKa = 10.24DD153 pKa = 3.57TMAILIIGLMFGSVTASVVGVLSYY177 pKa = 9.09FSPAEE182 pKa = 3.67QLQQYY187 pKa = 7.02VFWSFGSLGNLSWQALGVLTLCWLFGMILAIGRR220 pKa = 11.84IKK222 pKa = 10.73SLNSLLLGTRR232 pKa = 11.84YY233 pKa = 9.38AQSMGLRR240 pKa = 11.84VRR242 pKa = 11.84RR243 pKa = 11.84TTLLLIVATSLLAGGITAFAGPIAFVGLAVPHH275 pKa = 6.46LTRR278 pKa = 11.84QLVPSADD285 pKa = 3.35HH286 pKa = 6.94RR287 pKa = 11.84LLLPAVVLFGAALMLICDD305 pKa = 5.37TIAQLPGSEE314 pKa = 3.99FTLPINAITSLVGAPVVIWLLVRR337 pKa = 11.84KK338 pKa = 9.59RR339 pKa = 11.84KK340 pKa = 10.16LMFF343 pKa = 4.27
Molecular weight: 36.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
881571 |
38 |
4674 |
333.3 |
37.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.87 ± 0.043 | 0.711 ± 0.015 |
6.151 ± 0.052 | 6.647 ± 0.048 |
4.869 ± 0.04 | 7.212 ± 0.053 |
1.805 ± 0.027 | 6.985 ± 0.035 |
5.64 ± 0.062 | 9.634 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.476 ± 0.027 | 4.995 ± 0.041 |
3.825 ± 0.028 | 4.089 ± 0.029 |
4.574 ± 0.043 | 6.429 ± 0.037 |
5.365 ± 0.057 | 6.588 ± 0.045 |
1.232 ± 0.017 | 3.902 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
