
Lachnoclostridium sp. An298
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; unclassified Lachnoclostridium
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
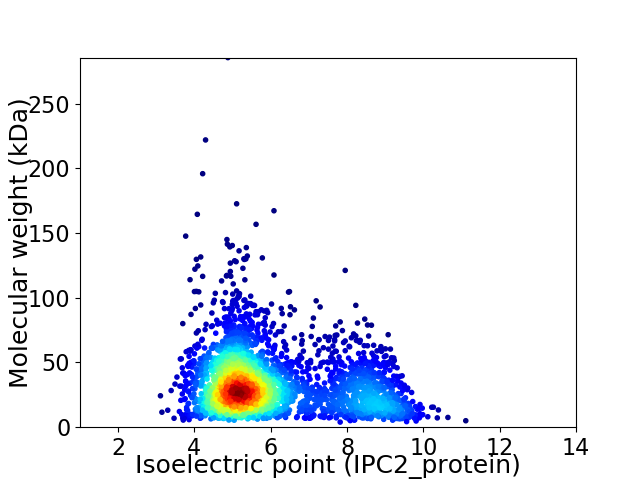
Virtual 2D-PAGE plot for 2948 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4B3Z5|A0A1Y4B3Z5_9FIRM Flavin reductase OS=Lachnoclostridium sp. An298 OX=1965627 GN=B5F86_10060 PE=4 SV=1
MM1 pKa = 7.83RR2 pKa = 11.84KK3 pKa = 9.52SKK5 pKa = 10.74LLICALFMSLILTALPGMEE24 pKa = 4.55SYY26 pKa = 11.11AAGEE30 pKa = 4.37TGEE33 pKa = 4.76SAAQTDD39 pKa = 4.13LQDD42 pKa = 3.65EE43 pKa = 4.32NGQRR47 pKa = 11.84LEE49 pKa = 4.34TVTAMDD55 pKa = 3.89EE56 pKa = 3.98NGNIYY61 pKa = 10.36EE62 pKa = 4.49VGDD65 pKa = 3.43SDD67 pKa = 4.04GTVGEE72 pKa = 4.62SAAEE76 pKa = 4.37DD77 pKa = 3.97GSEE80 pKa = 4.02EE81 pKa = 4.21GTDD84 pKa = 3.5AEE86 pKa = 4.61SGISLFSARR95 pKa = 11.84SAAVSMKK102 pKa = 10.08VVNFNTKK109 pKa = 10.12SNATTDD115 pKa = 3.86FTYY118 pKa = 10.71SGGSGYY124 pKa = 9.47TNGDD128 pKa = 3.35YY129 pKa = 11.2GADD132 pKa = 3.39AAYY135 pKa = 10.6LGTDD139 pKa = 3.39GSGDD143 pKa = 3.38IRR145 pKa = 11.84FMLAGVTGTVKK156 pKa = 10.35KK157 pKa = 10.93SEE159 pKa = 4.24VEE161 pKa = 4.15VVDD164 pKa = 3.97YY165 pKa = 11.45SSVADD170 pKa = 3.41NVSYY174 pKa = 11.44YY175 pKa = 10.19EE176 pKa = 3.96VSGGQLIHH184 pKa = 6.54YY185 pKa = 9.89ISYY188 pKa = 10.55NLNEE192 pKa = 4.88GPSSSINNGPAPSYY206 pKa = 10.31LSEE209 pKa = 3.88GVKK212 pKa = 10.03YY213 pKa = 10.51FSYY216 pKa = 10.45DD217 pKa = 2.58GHH219 pKa = 7.06YY220 pKa = 10.29FYY222 pKa = 11.28EE223 pKa = 4.91DD224 pKa = 3.89YY225 pKa = 10.04KK226 pKa = 11.7TMLNDD231 pKa = 3.51YY232 pKa = 10.74QSGSNGASAVNCGNAFYY249 pKa = 10.93NYY251 pKa = 10.01FQFLDD256 pKa = 3.89MNSSTNYY263 pKa = 10.63SGDD266 pKa = 3.6EE267 pKa = 3.96LDD269 pKa = 5.37SILSSKK275 pKa = 10.27ISSSSSKK282 pKa = 10.92LLGTGTLFVKK292 pKa = 9.97YY293 pKa = 8.71QNKK296 pKa = 9.83YY297 pKa = 9.8SVNALLSLGIAVNEE311 pKa = 4.51SAWGTSSICMDD322 pKa = 3.57KK323 pKa = 11.51NNLFGLNAVDD333 pKa = 3.94TSPGQSANYY342 pKa = 7.86FASVEE347 pKa = 4.04DD348 pKa = 4.6CIRR351 pKa = 11.84EE352 pKa = 4.03FMSEE356 pKa = 3.74WMADD360 pKa = 3.67GYY362 pKa = 11.15LSSSDD367 pKa = 2.97WRR369 pKa = 11.84NHH371 pKa = 6.07GEE373 pKa = 3.81LLGNKK378 pKa = 9.9GEE380 pKa = 4.83GINVSYY386 pKa = 11.09ASDD389 pKa = 4.16PYY391 pKa = 10.24WGEE394 pKa = 3.58KK395 pKa = 9.27AAAIAWEE402 pKa = 4.15LDD404 pKa = 3.46AAGGSHH410 pKa = 7.91DD411 pKa = 4.16YY412 pKa = 11.27LGISSGSGNEE422 pKa = 3.96SSEE425 pKa = 4.78DD426 pKa = 3.54NTDD429 pKa = 3.51TPSEE433 pKa = 4.3DD434 pKa = 3.43NMPPSDD440 pKa = 5.13AGGTDD445 pKa = 3.59DD446 pKa = 6.21ANDD449 pKa = 3.34SSAADD454 pKa = 3.81NDD456 pKa = 4.03SDD458 pKa = 3.85TDD460 pKa = 3.88DD461 pKa = 4.55PSEE464 pKa = 4.14EE465 pKa = 4.64DD466 pKa = 3.52SDD468 pKa = 4.79SEE470 pKa = 4.34ADD472 pKa = 3.64SSVDD476 pKa = 3.14TGNGSEE482 pKa = 4.55TEE484 pKa = 4.23NPTDD488 pKa = 3.69ANEE491 pKa = 4.35PSDD494 pKa = 4.0TNDD497 pKa = 3.15PTDD500 pKa = 3.68ANEE503 pKa = 4.47PSDD506 pKa = 4.08TNDD509 pKa = 3.2PTDD512 pKa = 3.48TNEE515 pKa = 4.24PSDD518 pKa = 3.35TGTPSEE524 pKa = 4.67EE525 pKa = 4.34EE526 pKa = 4.0NDD528 pKa = 3.8SEE530 pKa = 4.45TDD532 pKa = 3.32GGVDD536 pKa = 3.48EE537 pKa = 5.25EE538 pKa = 4.69NGSEE542 pKa = 5.09AEE544 pKa = 4.76DD545 pKa = 3.42PTDD548 pKa = 3.88ANEE551 pKa = 4.4PSDD554 pKa = 4.72ANDD557 pKa = 3.69PADD560 pKa = 3.74TNEE563 pKa = 5.1PIDD566 pKa = 3.69TNKK569 pKa = 10.28PVDD572 pKa = 3.58TDD574 pKa = 3.87TPSDD578 pKa = 3.56EE579 pKa = 4.59SSNSGADD586 pKa = 3.58KK587 pKa = 10.96KK588 pKa = 10.81PADD591 pKa = 4.04TPADD595 pKa = 3.99RR596 pKa = 11.84QLADD600 pKa = 3.28IDD602 pKa = 4.23HH603 pKa = 6.39GVSVSGRR610 pKa = 11.84FSEE613 pKa = 4.95DD614 pKa = 2.71AALNVQAVEE623 pKa = 4.32PNTDD627 pKa = 2.76QYY629 pKa = 10.79ATYY632 pKa = 9.26VTADD636 pKa = 3.57PVKK639 pKa = 9.42GKK641 pKa = 10.47VVLGVYY647 pKa = 10.31DD648 pKa = 3.44ITLTGTLEE656 pKa = 4.41GEE658 pKa = 4.09AQLTFRR664 pKa = 11.84VGTEE668 pKa = 3.68YY669 pKa = 10.89NGRR672 pKa = 11.84NVIILHH678 pKa = 5.59YY679 pKa = 10.25ADD681 pKa = 4.84DD682 pKa = 4.62GSYY685 pKa = 8.47EE686 pKa = 4.18TYY688 pKa = 10.35KK689 pKa = 11.01AAVEE693 pKa = 4.04NGQVTISVKK702 pKa = 10.36GFSPYY707 pKa = 10.53VIALDD712 pKa = 4.32DD713 pKa = 4.86ANSSSSIEE721 pKa = 3.94NKK723 pKa = 10.26APQTGDD729 pKa = 3.03TAEE732 pKa = 4.22PGIWIALMGMAIVLGAAAIFKK753 pKa = 10.16KK754 pKa = 10.39RR755 pKa = 11.84AVRR758 pKa = 3.75
MM1 pKa = 7.83RR2 pKa = 11.84KK3 pKa = 9.52SKK5 pKa = 10.74LLICALFMSLILTALPGMEE24 pKa = 4.55SYY26 pKa = 11.11AAGEE30 pKa = 4.37TGEE33 pKa = 4.76SAAQTDD39 pKa = 4.13LQDD42 pKa = 3.65EE43 pKa = 4.32NGQRR47 pKa = 11.84LEE49 pKa = 4.34TVTAMDD55 pKa = 3.89EE56 pKa = 3.98NGNIYY61 pKa = 10.36EE62 pKa = 4.49VGDD65 pKa = 3.43SDD67 pKa = 4.04GTVGEE72 pKa = 4.62SAAEE76 pKa = 4.37DD77 pKa = 3.97GSEE80 pKa = 4.02EE81 pKa = 4.21GTDD84 pKa = 3.5AEE86 pKa = 4.61SGISLFSARR95 pKa = 11.84SAAVSMKK102 pKa = 10.08VVNFNTKK109 pKa = 10.12SNATTDD115 pKa = 3.86FTYY118 pKa = 10.71SGGSGYY124 pKa = 9.47TNGDD128 pKa = 3.35YY129 pKa = 11.2GADD132 pKa = 3.39AAYY135 pKa = 10.6LGTDD139 pKa = 3.39GSGDD143 pKa = 3.38IRR145 pKa = 11.84FMLAGVTGTVKK156 pKa = 10.35KK157 pKa = 10.93SEE159 pKa = 4.24VEE161 pKa = 4.15VVDD164 pKa = 3.97YY165 pKa = 11.45SSVADD170 pKa = 3.41NVSYY174 pKa = 11.44YY175 pKa = 10.19EE176 pKa = 3.96VSGGQLIHH184 pKa = 6.54YY185 pKa = 9.89ISYY188 pKa = 10.55NLNEE192 pKa = 4.88GPSSSINNGPAPSYY206 pKa = 10.31LSEE209 pKa = 3.88GVKK212 pKa = 10.03YY213 pKa = 10.51FSYY216 pKa = 10.45DD217 pKa = 2.58GHH219 pKa = 7.06YY220 pKa = 10.29FYY222 pKa = 11.28EE223 pKa = 4.91DD224 pKa = 3.89YY225 pKa = 10.04KK226 pKa = 11.7TMLNDD231 pKa = 3.51YY232 pKa = 10.74QSGSNGASAVNCGNAFYY249 pKa = 10.93NYY251 pKa = 10.01FQFLDD256 pKa = 3.89MNSSTNYY263 pKa = 10.63SGDD266 pKa = 3.6EE267 pKa = 3.96LDD269 pKa = 5.37SILSSKK275 pKa = 10.27ISSSSSKK282 pKa = 10.92LLGTGTLFVKK292 pKa = 9.97YY293 pKa = 8.71QNKK296 pKa = 9.83YY297 pKa = 9.8SVNALLSLGIAVNEE311 pKa = 4.51SAWGTSSICMDD322 pKa = 3.57KK323 pKa = 11.51NNLFGLNAVDD333 pKa = 3.94TSPGQSANYY342 pKa = 7.86FASVEE347 pKa = 4.04DD348 pKa = 4.6CIRR351 pKa = 11.84EE352 pKa = 4.03FMSEE356 pKa = 3.74WMADD360 pKa = 3.67GYY362 pKa = 11.15LSSSDD367 pKa = 2.97WRR369 pKa = 11.84NHH371 pKa = 6.07GEE373 pKa = 3.81LLGNKK378 pKa = 9.9GEE380 pKa = 4.83GINVSYY386 pKa = 11.09ASDD389 pKa = 4.16PYY391 pKa = 10.24WGEE394 pKa = 3.58KK395 pKa = 9.27AAAIAWEE402 pKa = 4.15LDD404 pKa = 3.46AAGGSHH410 pKa = 7.91DD411 pKa = 4.16YY412 pKa = 11.27LGISSGSGNEE422 pKa = 3.96SSEE425 pKa = 4.78DD426 pKa = 3.54NTDD429 pKa = 3.51TPSEE433 pKa = 4.3DD434 pKa = 3.43NMPPSDD440 pKa = 5.13AGGTDD445 pKa = 3.59DD446 pKa = 6.21ANDD449 pKa = 3.34SSAADD454 pKa = 3.81NDD456 pKa = 4.03SDD458 pKa = 3.85TDD460 pKa = 3.88DD461 pKa = 4.55PSEE464 pKa = 4.14EE465 pKa = 4.64DD466 pKa = 3.52SDD468 pKa = 4.79SEE470 pKa = 4.34ADD472 pKa = 3.64SSVDD476 pKa = 3.14TGNGSEE482 pKa = 4.55TEE484 pKa = 4.23NPTDD488 pKa = 3.69ANEE491 pKa = 4.35PSDD494 pKa = 4.0TNDD497 pKa = 3.15PTDD500 pKa = 3.68ANEE503 pKa = 4.47PSDD506 pKa = 4.08TNDD509 pKa = 3.2PTDD512 pKa = 3.48TNEE515 pKa = 4.24PSDD518 pKa = 3.35TGTPSEE524 pKa = 4.67EE525 pKa = 4.34EE526 pKa = 4.0NDD528 pKa = 3.8SEE530 pKa = 4.45TDD532 pKa = 3.32GGVDD536 pKa = 3.48EE537 pKa = 5.25EE538 pKa = 4.69NGSEE542 pKa = 5.09AEE544 pKa = 4.76DD545 pKa = 3.42PTDD548 pKa = 3.88ANEE551 pKa = 4.4PSDD554 pKa = 4.72ANDD557 pKa = 3.69PADD560 pKa = 3.74TNEE563 pKa = 5.1PIDD566 pKa = 3.69TNKK569 pKa = 10.28PVDD572 pKa = 3.58TDD574 pKa = 3.87TPSDD578 pKa = 3.56EE579 pKa = 4.59SSNSGADD586 pKa = 3.58KK587 pKa = 10.96KK588 pKa = 10.81PADD591 pKa = 4.04TPADD595 pKa = 3.99RR596 pKa = 11.84QLADD600 pKa = 3.28IDD602 pKa = 4.23HH603 pKa = 6.39GVSVSGRR610 pKa = 11.84FSEE613 pKa = 4.95DD614 pKa = 2.71AALNVQAVEE623 pKa = 4.32PNTDD627 pKa = 2.76QYY629 pKa = 10.79ATYY632 pKa = 9.26VTADD636 pKa = 3.57PVKK639 pKa = 9.42GKK641 pKa = 10.47VVLGVYY647 pKa = 10.31DD648 pKa = 3.44ITLTGTLEE656 pKa = 4.41GEE658 pKa = 4.09AQLTFRR664 pKa = 11.84VGTEE668 pKa = 3.68YY669 pKa = 10.89NGRR672 pKa = 11.84NVIILHH678 pKa = 5.59YY679 pKa = 10.25ADD681 pKa = 4.84DD682 pKa = 4.62GSYY685 pKa = 8.47EE686 pKa = 4.18TYY688 pKa = 10.35KK689 pKa = 11.01AAVEE693 pKa = 4.04NGQVTISVKK702 pKa = 10.36GFSPYY707 pKa = 10.53VIALDD712 pKa = 4.32DD713 pKa = 4.86ANSSSSIEE721 pKa = 3.94NKK723 pKa = 10.26APQTGDD729 pKa = 3.03TAEE732 pKa = 4.22PGIWIALMGMAIVLGAAAIFKK753 pKa = 10.16KK754 pKa = 10.39RR755 pKa = 11.84AVRR758 pKa = 3.75
Molecular weight: 80.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4AQ20|A0A1Y4AQ20_9FIRM Uncharacterized protein OS=Lachnoclostridium sp. An298 OX=1965627 GN=B5F86_15180 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTPGGRR28 pKa = 11.84KK29 pKa = 8.8VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84KK40 pKa = 8.18QLSAA44 pKa = 3.9
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
916666 |
36 |
2524 |
310.9 |
34.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.644 ± 0.045 | 1.52 ± 0.02 |
5.928 ± 0.04 | 7.892 ± 0.049 |
3.941 ± 0.035 | 7.294 ± 0.039 |
1.749 ± 0.02 | 7.278 ± 0.043 |
6.282 ± 0.043 | 8.781 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.128 ± 0.023 | 3.954 ± 0.026 |
3.411 ± 0.026 | 3.143 ± 0.025 |
5.035 ± 0.035 | 5.698 ± 0.036 |
5.355 ± 0.029 | 6.871 ± 0.038 |
0.922 ± 0.017 | 4.172 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
