
Suakwa aphid-borne yellows virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
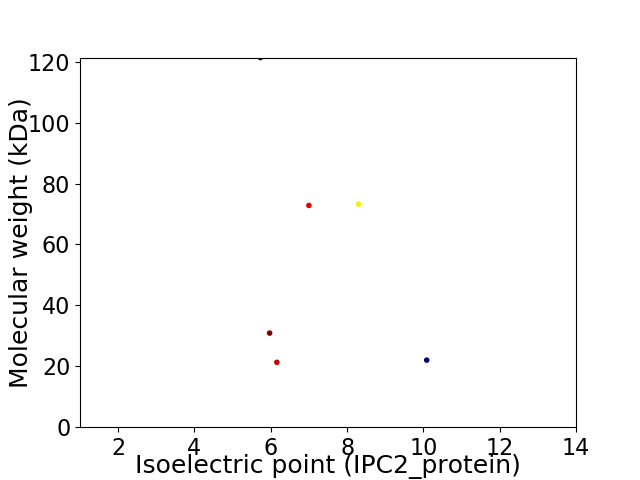
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7FDA2|J7FDA2_9LUTE Movement protein OS=Suakwa aphid-borne yellows virus OX=646010 PE=3 SV=1
MM1 pKa = 7.68KK2 pKa = 10.62NMQFVVLFFFCLSASTLCWPEE23 pKa = 3.86VEE25 pKa = 4.72PEE27 pKa = 4.15SAQALYY33 pKa = 9.89PGPTYY38 pKa = 10.82NLSYY42 pKa = 11.02DD43 pKa = 3.7GLSVLEE49 pKa = 4.95LPFHH53 pKa = 6.42TDD55 pKa = 2.09WKK57 pKa = 10.09EE58 pKa = 3.58IKK60 pKa = 10.12EE61 pKa = 4.1NFGFFCEE68 pKa = 4.56EE69 pKa = 4.19PGEE72 pKa = 4.3LDD74 pKa = 3.74LNQLIAEE81 pKa = 4.33SSYY84 pKa = 11.51TDD86 pKa = 2.99ILTSLSNKK94 pKa = 9.46ILLDD98 pKa = 3.99FKK100 pKa = 11.18SVVSVALDD108 pKa = 3.36TFLVFWALCKK118 pKa = 10.78GSIQAYY124 pKa = 9.64AFRR127 pKa = 11.84AFKK130 pKa = 10.52AWLEE134 pKa = 4.16VIITLWISVISAILSGLTWALLNIFPHH161 pKa = 5.79TLTLIFLFFFTNLLVKK177 pKa = 9.83GFRR180 pKa = 11.84ILCGTACLAFLRR192 pKa = 11.84MLFSPLLPVKK202 pKa = 10.54RR203 pKa = 11.84ILTWTLARR211 pKa = 11.84LLGNSFGRR219 pKa = 11.84KK220 pKa = 8.65LEE222 pKa = 4.11KK223 pKa = 9.99ATEE226 pKa = 4.01GWKK229 pKa = 10.61SFEE232 pKa = 4.35IKK234 pKa = 9.22MAPPSSSALPVRR246 pKa = 11.84NRR248 pKa = 11.84ITKK251 pKa = 8.99EE252 pKa = 3.38HH253 pKa = 6.61AGYY256 pKa = 10.49CGFVQTHH263 pKa = 5.9MKK265 pKa = 10.1NADD268 pKa = 3.23GTYY271 pKa = 9.78VEE273 pKa = 4.89AFITARR279 pKa = 11.84HH280 pKa = 6.03VIDD283 pKa = 5.74DD284 pKa = 4.36DD285 pKa = 4.02LGEE288 pKa = 4.04IHH290 pKa = 7.15SPKK293 pKa = 9.44TKK295 pKa = 10.85NFIACADD302 pKa = 4.07LEE304 pKa = 4.41VLGDD308 pKa = 3.69WEE310 pKa = 5.03SIDD313 pKa = 5.9LILLRR318 pKa = 11.84APKK321 pKa = 9.79HH322 pKa = 4.88ARR324 pKa = 11.84SYY326 pKa = 10.63LGCKK330 pKa = 8.13PASMVAIDD338 pKa = 5.47RR339 pKa = 11.84LATCDD344 pKa = 3.25ASTFHH349 pKa = 7.27LIDD352 pKa = 4.75GEE354 pKa = 4.27WHH356 pKa = 5.96MSNARR361 pKa = 11.84LVGDD365 pKa = 4.0VKK367 pKa = 11.26NGVAVLSNSVCGHH380 pKa = 6.13SGSPFYY386 pKa = 10.59NGKK389 pKa = 9.8HH390 pKa = 5.36ILGVLYY396 pKa = 10.86GGGSYY401 pKa = 10.73FEE403 pKa = 5.09NFNVMNPIPAIPGLTTPEE421 pKa = 3.74RR422 pKa = 11.84RR423 pKa = 11.84LEE425 pKa = 4.08TTAPRR430 pKa = 11.84GKK432 pKa = 10.02IFNNEE437 pKa = 3.67DD438 pKa = 3.22FFQKK442 pKa = 10.79LEE444 pKa = 4.01EE445 pKa = 5.07EE446 pKa = 4.36YY447 pKa = 11.17LQMKK451 pKa = 9.03NFKK454 pKa = 10.58SKK456 pKa = 9.56TGVDD460 pKa = 3.43WADD463 pKa = 3.69LPSDD467 pKa = 4.92DD468 pKa = 5.86EE469 pKa = 4.77MDD471 pKa = 4.01FQDD474 pKa = 4.53ALPSFRR480 pKa = 11.84KK481 pKa = 9.84AEE483 pKa = 4.09KK484 pKa = 10.43AEE486 pKa = 4.2PSHH489 pKa = 6.61PVANTRR495 pKa = 11.84EE496 pKa = 4.08PAVVVGDD503 pKa = 3.63IRR505 pKa = 11.84CNISGNRR512 pKa = 11.84RR513 pKa = 11.84EE514 pKa = 4.12QGRR517 pKa = 11.84LLQQRR522 pKa = 11.84PHH524 pKa = 5.64VRR526 pKa = 11.84CCEE529 pKa = 4.03EE530 pKa = 3.87PRR532 pKa = 11.84EE533 pKa = 4.31SCGPDD538 pKa = 2.98GGQCGHH544 pKa = 7.37PGDD547 pKa = 4.09PTRR550 pKa = 11.84GSSDD554 pKa = 3.37SCCPALRR561 pKa = 11.84QPEE564 pKa = 4.19KK565 pKa = 10.91DD566 pKa = 3.28EE567 pKa = 4.42SRR569 pKa = 11.84TVPAWEE575 pKa = 4.23SRR577 pKa = 11.84GTEE580 pKa = 3.29ADD582 pKa = 2.76NFRR585 pKa = 11.84RR586 pKa = 11.84YY587 pKa = 8.43FQNEE591 pKa = 3.81YY592 pKa = 9.83DD593 pKa = 3.58WEE595 pKa = 4.38VRR597 pKa = 11.84APAPQVPGFRR607 pKa = 11.84HH608 pKa = 6.55CGTLPRR614 pKa = 11.84YY615 pKa = 9.32YY616 pKa = 10.67FSRR619 pKa = 11.84QKK621 pKa = 11.4GEE623 pKa = 4.26TEE625 pKa = 3.39WGKK628 pKa = 7.87TLTRR632 pKa = 11.84DD633 pKa = 3.51NPSLGEE639 pKa = 3.88ATAGYY644 pKa = 8.93GWPEE648 pKa = 4.22FGPGAEE654 pKa = 4.26LKK656 pKa = 10.47SLKK659 pKa = 10.2LQAARR664 pKa = 11.84WRR666 pKa = 11.84EE667 pKa = 3.81RR668 pKa = 11.84ASSAKK673 pKa = 9.91VPYY676 pKa = 8.82PSEE679 pKa = 4.07RR680 pKa = 11.84EE681 pKa = 3.76RR682 pKa = 11.84VIRR685 pKa = 11.84IATEE689 pKa = 4.12RR690 pKa = 11.84YY691 pKa = 9.28SKK693 pKa = 10.77VKK695 pKa = 10.42SVAPACSRR703 pKa = 11.84SGEE706 pKa = 4.28LKK708 pKa = 10.16WEE710 pKa = 4.15GLVEE714 pKa = 4.21DD715 pKa = 5.59LKK717 pKa = 11.53DD718 pKa = 3.96CLPSLSLEE726 pKa = 4.37AGVGVPYY733 pKa = 10.23IAYY736 pKa = 9.16GRR738 pKa = 11.84PTLGSWVNDD747 pKa = 3.36QALLPILSRR756 pKa = 11.84LVFDD760 pKa = 4.62RR761 pKa = 11.84LKK763 pKa = 11.13KK764 pKa = 9.41MSEE767 pKa = 3.83TRR769 pKa = 11.84FDD771 pKa = 4.17DD772 pKa = 3.59MTPEE776 pKa = 3.95EE777 pKa = 4.43LVQNGLCDD785 pKa = 4.91PIRR788 pKa = 11.84TFIKK792 pKa = 10.71GEE794 pKa = 3.74PHH796 pKa = 6.74KK797 pKa = 10.88SSKK800 pKa = 10.55LKK802 pKa = 9.58EE803 pKa = 3.75GRR805 pKa = 11.84YY806 pKa = 9.15RR807 pKa = 11.84LIMSVSIVDD816 pKa = 3.48QLVARR821 pKa = 11.84VLFQNQNKK829 pKa = 10.05SEE831 pKa = 4.05IALWRR836 pKa = 11.84AIPSKK841 pKa = 10.48PGLGLSTDD849 pKa = 3.62EE850 pKa = 3.99QVLDD854 pKa = 4.63FVHH857 pKa = 6.47NLAEE861 pKa = 3.99NTEE864 pKa = 4.11YY865 pKa = 10.84RR866 pKa = 11.84EE867 pKa = 4.15SVEE870 pKa = 5.28DD871 pKa = 5.81FIKK874 pKa = 9.87HH875 pKa = 3.86WHH877 pKa = 5.55RR878 pKa = 11.84TVVPTDD884 pKa = 3.24CSGFDD889 pKa = 3.34WSVSSWMLEE898 pKa = 3.48DD899 pKa = 4.98DD900 pKa = 3.64MEE902 pKa = 4.34VRR904 pKa = 11.84NNLTINNNEE913 pKa = 3.64LTRR916 pKa = 11.84RR917 pKa = 11.84LRR919 pKa = 11.84ACWLKK924 pKa = 10.87CISNSVICLSDD935 pKa = 3.39GTLLAQEE942 pKa = 4.58FPGIQKK948 pKa = 10.23SGSYY952 pKa = 8.29NTSSTNSRR960 pKa = 11.84VRR962 pKa = 11.84VMLSIYY968 pKa = 10.65AGASWAIAMGDD979 pKa = 3.83DD980 pKa = 3.85ALEE983 pKa = 4.36SADD986 pKa = 4.21TIIPLYY992 pKa = 9.06EE993 pKa = 3.85QYY995 pKa = 11.12GLKK998 pKa = 10.38VEE1000 pKa = 4.48VSDD1003 pKa = 3.87KK1004 pKa = 11.67LEE1006 pKa = 4.38FCSHH1010 pKa = 5.84EE1011 pKa = 4.46FKK1013 pKa = 10.94SSSLAIPTNIGKK1025 pKa = 8.31MLYY1028 pKa = 10.21KK1029 pKa = 10.39LIHH1032 pKa = 6.61GYY1034 pKa = 10.48EE1035 pKa = 4.06IFPGMSSIQVTINYY1049 pKa = 7.63LTACYY1054 pKa = 10.21SVLNEE1059 pKa = 4.0LRR1061 pKa = 11.84HH1062 pKa = 5.29VPEE1065 pKa = 4.4AASLVAQCLAPVEE1078 pKa = 4.18AQKK1081 pKa = 11.17VV1082 pKa = 3.51
MM1 pKa = 7.68KK2 pKa = 10.62NMQFVVLFFFCLSASTLCWPEE23 pKa = 3.86VEE25 pKa = 4.72PEE27 pKa = 4.15SAQALYY33 pKa = 9.89PGPTYY38 pKa = 10.82NLSYY42 pKa = 11.02DD43 pKa = 3.7GLSVLEE49 pKa = 4.95LPFHH53 pKa = 6.42TDD55 pKa = 2.09WKK57 pKa = 10.09EE58 pKa = 3.58IKK60 pKa = 10.12EE61 pKa = 4.1NFGFFCEE68 pKa = 4.56EE69 pKa = 4.19PGEE72 pKa = 4.3LDD74 pKa = 3.74LNQLIAEE81 pKa = 4.33SSYY84 pKa = 11.51TDD86 pKa = 2.99ILTSLSNKK94 pKa = 9.46ILLDD98 pKa = 3.99FKK100 pKa = 11.18SVVSVALDD108 pKa = 3.36TFLVFWALCKK118 pKa = 10.78GSIQAYY124 pKa = 9.64AFRR127 pKa = 11.84AFKK130 pKa = 10.52AWLEE134 pKa = 4.16VIITLWISVISAILSGLTWALLNIFPHH161 pKa = 5.79TLTLIFLFFFTNLLVKK177 pKa = 9.83GFRR180 pKa = 11.84ILCGTACLAFLRR192 pKa = 11.84MLFSPLLPVKK202 pKa = 10.54RR203 pKa = 11.84ILTWTLARR211 pKa = 11.84LLGNSFGRR219 pKa = 11.84KK220 pKa = 8.65LEE222 pKa = 4.11KK223 pKa = 9.99ATEE226 pKa = 4.01GWKK229 pKa = 10.61SFEE232 pKa = 4.35IKK234 pKa = 9.22MAPPSSSALPVRR246 pKa = 11.84NRR248 pKa = 11.84ITKK251 pKa = 8.99EE252 pKa = 3.38HH253 pKa = 6.61AGYY256 pKa = 10.49CGFVQTHH263 pKa = 5.9MKK265 pKa = 10.1NADD268 pKa = 3.23GTYY271 pKa = 9.78VEE273 pKa = 4.89AFITARR279 pKa = 11.84HH280 pKa = 6.03VIDD283 pKa = 5.74DD284 pKa = 4.36DD285 pKa = 4.02LGEE288 pKa = 4.04IHH290 pKa = 7.15SPKK293 pKa = 9.44TKK295 pKa = 10.85NFIACADD302 pKa = 4.07LEE304 pKa = 4.41VLGDD308 pKa = 3.69WEE310 pKa = 5.03SIDD313 pKa = 5.9LILLRR318 pKa = 11.84APKK321 pKa = 9.79HH322 pKa = 4.88ARR324 pKa = 11.84SYY326 pKa = 10.63LGCKK330 pKa = 8.13PASMVAIDD338 pKa = 5.47RR339 pKa = 11.84LATCDD344 pKa = 3.25ASTFHH349 pKa = 7.27LIDD352 pKa = 4.75GEE354 pKa = 4.27WHH356 pKa = 5.96MSNARR361 pKa = 11.84LVGDD365 pKa = 4.0VKK367 pKa = 11.26NGVAVLSNSVCGHH380 pKa = 6.13SGSPFYY386 pKa = 10.59NGKK389 pKa = 9.8HH390 pKa = 5.36ILGVLYY396 pKa = 10.86GGGSYY401 pKa = 10.73FEE403 pKa = 5.09NFNVMNPIPAIPGLTTPEE421 pKa = 3.74RR422 pKa = 11.84RR423 pKa = 11.84LEE425 pKa = 4.08TTAPRR430 pKa = 11.84GKK432 pKa = 10.02IFNNEE437 pKa = 3.67DD438 pKa = 3.22FFQKK442 pKa = 10.79LEE444 pKa = 4.01EE445 pKa = 5.07EE446 pKa = 4.36YY447 pKa = 11.17LQMKK451 pKa = 9.03NFKK454 pKa = 10.58SKK456 pKa = 9.56TGVDD460 pKa = 3.43WADD463 pKa = 3.69LPSDD467 pKa = 4.92DD468 pKa = 5.86EE469 pKa = 4.77MDD471 pKa = 4.01FQDD474 pKa = 4.53ALPSFRR480 pKa = 11.84KK481 pKa = 9.84AEE483 pKa = 4.09KK484 pKa = 10.43AEE486 pKa = 4.2PSHH489 pKa = 6.61PVANTRR495 pKa = 11.84EE496 pKa = 4.08PAVVVGDD503 pKa = 3.63IRR505 pKa = 11.84CNISGNRR512 pKa = 11.84RR513 pKa = 11.84EE514 pKa = 4.12QGRR517 pKa = 11.84LLQQRR522 pKa = 11.84PHH524 pKa = 5.64VRR526 pKa = 11.84CCEE529 pKa = 4.03EE530 pKa = 3.87PRR532 pKa = 11.84EE533 pKa = 4.31SCGPDD538 pKa = 2.98GGQCGHH544 pKa = 7.37PGDD547 pKa = 4.09PTRR550 pKa = 11.84GSSDD554 pKa = 3.37SCCPALRR561 pKa = 11.84QPEE564 pKa = 4.19KK565 pKa = 10.91DD566 pKa = 3.28EE567 pKa = 4.42SRR569 pKa = 11.84TVPAWEE575 pKa = 4.23SRR577 pKa = 11.84GTEE580 pKa = 3.29ADD582 pKa = 2.76NFRR585 pKa = 11.84RR586 pKa = 11.84YY587 pKa = 8.43FQNEE591 pKa = 3.81YY592 pKa = 9.83DD593 pKa = 3.58WEE595 pKa = 4.38VRR597 pKa = 11.84APAPQVPGFRR607 pKa = 11.84HH608 pKa = 6.55CGTLPRR614 pKa = 11.84YY615 pKa = 9.32YY616 pKa = 10.67FSRR619 pKa = 11.84QKK621 pKa = 11.4GEE623 pKa = 4.26TEE625 pKa = 3.39WGKK628 pKa = 7.87TLTRR632 pKa = 11.84DD633 pKa = 3.51NPSLGEE639 pKa = 3.88ATAGYY644 pKa = 8.93GWPEE648 pKa = 4.22FGPGAEE654 pKa = 4.26LKK656 pKa = 10.47SLKK659 pKa = 10.2LQAARR664 pKa = 11.84WRR666 pKa = 11.84EE667 pKa = 3.81RR668 pKa = 11.84ASSAKK673 pKa = 9.91VPYY676 pKa = 8.82PSEE679 pKa = 4.07RR680 pKa = 11.84EE681 pKa = 3.76RR682 pKa = 11.84VIRR685 pKa = 11.84IATEE689 pKa = 4.12RR690 pKa = 11.84YY691 pKa = 9.28SKK693 pKa = 10.77VKK695 pKa = 10.42SVAPACSRR703 pKa = 11.84SGEE706 pKa = 4.28LKK708 pKa = 10.16WEE710 pKa = 4.15GLVEE714 pKa = 4.21DD715 pKa = 5.59LKK717 pKa = 11.53DD718 pKa = 3.96CLPSLSLEE726 pKa = 4.37AGVGVPYY733 pKa = 10.23IAYY736 pKa = 9.16GRR738 pKa = 11.84PTLGSWVNDD747 pKa = 3.36QALLPILSRR756 pKa = 11.84LVFDD760 pKa = 4.62RR761 pKa = 11.84LKK763 pKa = 11.13KK764 pKa = 9.41MSEE767 pKa = 3.83TRR769 pKa = 11.84FDD771 pKa = 4.17DD772 pKa = 3.59MTPEE776 pKa = 3.95EE777 pKa = 4.43LVQNGLCDD785 pKa = 4.91PIRR788 pKa = 11.84TFIKK792 pKa = 10.71GEE794 pKa = 3.74PHH796 pKa = 6.74KK797 pKa = 10.88SSKK800 pKa = 10.55LKK802 pKa = 9.58EE803 pKa = 3.75GRR805 pKa = 11.84YY806 pKa = 9.15RR807 pKa = 11.84LIMSVSIVDD816 pKa = 3.48QLVARR821 pKa = 11.84VLFQNQNKK829 pKa = 10.05SEE831 pKa = 4.05IALWRR836 pKa = 11.84AIPSKK841 pKa = 10.48PGLGLSTDD849 pKa = 3.62EE850 pKa = 3.99QVLDD854 pKa = 4.63FVHH857 pKa = 6.47NLAEE861 pKa = 3.99NTEE864 pKa = 4.11YY865 pKa = 10.84RR866 pKa = 11.84EE867 pKa = 4.15SVEE870 pKa = 5.28DD871 pKa = 5.81FIKK874 pKa = 9.87HH875 pKa = 3.86WHH877 pKa = 5.55RR878 pKa = 11.84TVVPTDD884 pKa = 3.24CSGFDD889 pKa = 3.34WSVSSWMLEE898 pKa = 3.48DD899 pKa = 4.98DD900 pKa = 3.64MEE902 pKa = 4.34VRR904 pKa = 11.84NNLTINNNEE913 pKa = 3.64LTRR916 pKa = 11.84RR917 pKa = 11.84LRR919 pKa = 11.84ACWLKK924 pKa = 10.87CISNSVICLSDD935 pKa = 3.39GTLLAQEE942 pKa = 4.58FPGIQKK948 pKa = 10.23SGSYY952 pKa = 8.29NTSSTNSRR960 pKa = 11.84VRR962 pKa = 11.84VMLSIYY968 pKa = 10.65AGASWAIAMGDD979 pKa = 3.83DD980 pKa = 3.85ALEE983 pKa = 4.36SADD986 pKa = 4.21TIIPLYY992 pKa = 9.06EE993 pKa = 3.85QYY995 pKa = 11.12GLKK998 pKa = 10.38VEE1000 pKa = 4.48VSDD1003 pKa = 3.87KK1004 pKa = 11.67LEE1006 pKa = 4.38FCSHH1010 pKa = 5.84EE1011 pKa = 4.46FKK1013 pKa = 10.94SSSLAIPTNIGKK1025 pKa = 8.31MLYY1028 pKa = 10.21KK1029 pKa = 10.39LIHH1032 pKa = 6.61GYY1034 pKa = 10.48EE1035 pKa = 4.06IFPGMSSIQVTINYY1049 pKa = 7.63LTACYY1054 pKa = 10.21SVLNEE1059 pKa = 4.0LRR1061 pKa = 11.84HH1062 pKa = 5.29VPEE1065 pKa = 4.4AASLVAQCLAPVEE1078 pKa = 4.18AQKK1081 pKa = 11.17VV1082 pKa = 3.51
Molecular weight: 121.37 kDa
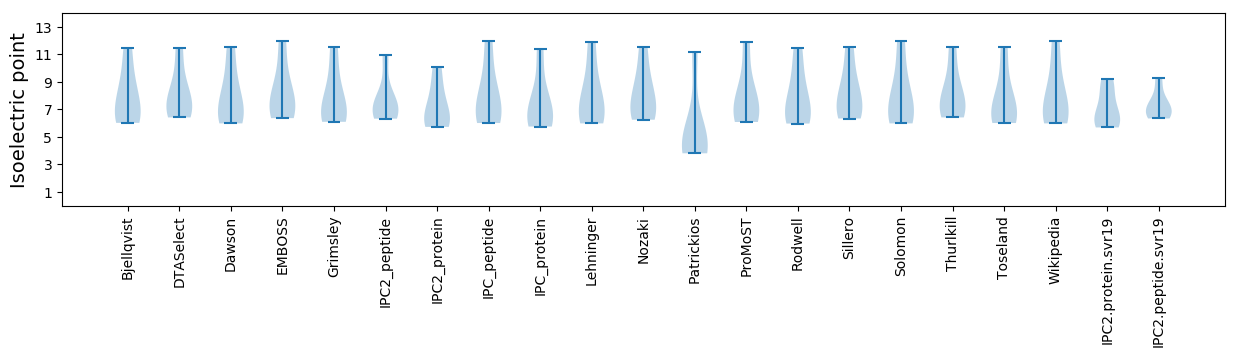
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7FE76|J7FE76_9LUTE p3 OS=Suakwa aphid-borne yellows virus OX=646010 PE=4 SV=1
MM1 pKa = 6.53NTVVVRR7 pKa = 11.84NRR9 pKa = 11.84NGMGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84NSRR20 pKa = 11.84RR21 pKa = 11.84NAKK24 pKa = 9.19GNRR27 pKa = 11.84VVVVQAPGAPQRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NGRR48 pKa = 11.84RR49 pKa = 11.84PSRR52 pKa = 11.84GGRR55 pKa = 11.84ARR57 pKa = 11.84GTSQGEE63 pKa = 4.12TFVFSKK69 pKa = 11.24DD70 pKa = 3.3NLTGSSSGAITFGPSLSDD88 pKa = 3.25SPAFSSGILKK98 pKa = 10.26AYY100 pKa = 10.18HH101 pKa = 6.62EE102 pKa = 4.74YY103 pKa = 10.05KK104 pKa = 10.27ISMVKK109 pKa = 10.66LEE111 pKa = 5.07FISEE115 pKa = 4.18ASSTSSGSISYY126 pKa = 10.24EE127 pKa = 3.75LDD129 pKa = 3.23PHH131 pKa = 6.86CKK133 pKa = 10.08LSSLQSTVNKK143 pKa = 10.33FGITKK148 pKa = 10.09SGSRR152 pKa = 11.84TWSAKK157 pKa = 9.97FINGMEE163 pKa = 3.99WHH165 pKa = 7.45DD166 pKa = 3.56ATEE169 pKa = 4.04DD170 pKa = 3.41QFRR173 pKa = 11.84ILYY176 pKa = 9.14KK177 pKa = 11.03GNGSSSTAGSFRR189 pKa = 11.84ITIRR193 pKa = 11.84CQVQNPKK200 pKa = 10.62
MM1 pKa = 6.53NTVVVRR7 pKa = 11.84NRR9 pKa = 11.84NGMGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84NSRR20 pKa = 11.84RR21 pKa = 11.84NAKK24 pKa = 9.19GNRR27 pKa = 11.84VVVVQAPGAPQRR39 pKa = 11.84GRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84NGRR48 pKa = 11.84RR49 pKa = 11.84PSRR52 pKa = 11.84GGRR55 pKa = 11.84ARR57 pKa = 11.84GTSQGEE63 pKa = 4.12TFVFSKK69 pKa = 11.24DD70 pKa = 3.3NLTGSSSGAITFGPSLSDD88 pKa = 3.25SPAFSSGILKK98 pKa = 10.26AYY100 pKa = 10.18HH101 pKa = 6.62EE102 pKa = 4.74YY103 pKa = 10.05KK104 pKa = 10.27ISMVKK109 pKa = 10.66LEE111 pKa = 5.07FISEE115 pKa = 4.18ASSTSSGSISYY126 pKa = 10.24EE127 pKa = 3.75LDD129 pKa = 3.23PHH131 pKa = 6.86CKK133 pKa = 10.08LSSLQSTVNKK143 pKa = 10.33FGITKK148 pKa = 10.09SGSRR152 pKa = 11.84TWSAKK157 pKa = 9.97FINGMEE163 pKa = 3.99WHH165 pKa = 7.45DD166 pKa = 3.56ATEE169 pKa = 4.04DD170 pKa = 3.41QFRR173 pKa = 11.84ILYY176 pKa = 9.14KK177 pKa = 11.03GNGSSSTAGSFRR189 pKa = 11.84ITIRR193 pKa = 11.84CQVQNPKK200 pKa = 10.62
Molecular weight: 22.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3062 |
192 |
1082 |
510.3 |
56.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.218 ± 0.499 | 2.025 ± 0.393 |
4.605 ± 0.242 | 6.434 ± 0.392 |
4.474 ± 0.582 | 7.185 ± 0.679 |
2.057 ± 0.094 | 4.931 ± 0.203 |
4.899 ± 0.415 | 9.014 ± 1.251 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.731 ± 0.1 | 4.442 ± 0.227 |
5.944 ± 0.414 | 3.135 ± 0.328 |
6.793 ± 0.72 | 9.242 ± 0.936 |
5.519 ± 0.272 | 5.487 ± 0.307 |
1.992 ± 0.157 | 2.841 ± 0.194 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
