
Martelella sp. BGMRC2036
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Martelella
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
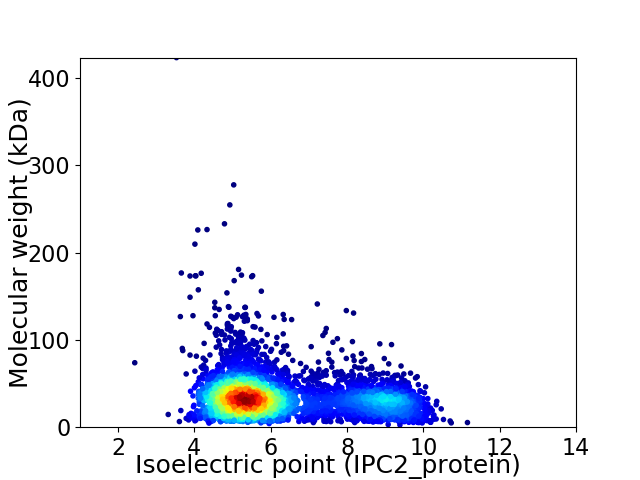
Virtual 2D-PAGE plot for 4394 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A506U505|A0A506U505_9RHIZ Cytochrome bo(3) ubiquinol oxidase subunit 4 OS=Martelella sp. BGMRC2036 OX=2590451 GN=cyoD PE=3 SV=1
MM1 pKa = 7.11LQRR4 pKa = 11.84RR5 pKa = 11.84EE6 pKa = 4.11LRR8 pKa = 11.84LLPPSLSISKK18 pKa = 10.72SMTGPHH24 pKa = 6.62LNLEE28 pKa = 4.25YY29 pKa = 10.35RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 11.01DD34 pKa = 4.17YY35 pKa = 11.33EE36 pKa = 4.24DD37 pKa = 3.6QPEE40 pKa = 4.43DD41 pKa = 3.72KK42 pKa = 10.98SIDD45 pKa = 3.17RR46 pKa = 11.84GTRR49 pKa = 11.84DD50 pKa = 3.2WIAAEE55 pKa = 4.01QPKK58 pKa = 10.11VYY60 pKa = 10.53HH61 pKa = 6.84FIFTVGDD68 pKa = 3.54DD69 pKa = 3.59RR70 pKa = 11.84LKK72 pKa = 10.34VTGTDD77 pKa = 3.45FDD79 pKa = 4.15TGFLADD85 pKa = 4.02MRR87 pKa = 11.84SGNDD91 pKa = 4.97FVTTWDD97 pKa = 3.72NPYY100 pKa = 10.8LPASTFWMGSGNDD113 pKa = 3.49VANVCGSSPTSDD125 pKa = 3.2EE126 pKa = 5.2DD127 pKa = 3.45EE128 pKa = 5.1DD129 pKa = 4.62EE130 pKa = 4.17EE131 pKa = 4.58VTRR134 pKa = 11.84VLCDD138 pKa = 4.51DD139 pKa = 4.95GDD141 pKa = 5.0DD142 pKa = 4.88GDD144 pKa = 5.75DD145 pKa = 4.37GDD147 pKa = 5.94DD148 pKa = 3.4IVNITGGGYY157 pKa = 11.0ANVFGDD163 pKa = 4.57DD164 pKa = 4.56GNDD167 pKa = 3.7TLCVQQNGAFKK178 pKa = 10.78SGVISLTGGAGYY190 pKa = 8.14DD191 pKa = 3.5TFLVTQHH198 pKa = 6.84PSSSYY203 pKa = 11.07NLPTSTGPASIGIFVTDD220 pKa = 3.12VLQDD224 pKa = 3.47VLGWVPGVGLATHH237 pKa = 7.18AAAALISTTTMAARR251 pKa = 11.84QSTRR255 pKa = 11.84FLNEE259 pKa = 3.15FSGAMDD265 pKa = 4.03GQDD268 pKa = 2.83GSVNIPQGGDD278 pKa = 3.17TYY280 pKa = 11.78VSIEE284 pKa = 4.47DD285 pKa = 3.92FDD287 pKa = 4.39PLEE290 pKa = 4.52DD291 pKa = 4.25QLVQTYY297 pKa = 8.29LHH299 pKa = 6.08NTNGMEE305 pKa = 4.06QYY307 pKa = 10.14SWQSQDD313 pKa = 3.17VASDD317 pKa = 3.38NFSIAFKK324 pKa = 10.98SDD326 pKa = 3.01GTNVSSVIHH335 pKa = 6.12VDD337 pKa = 3.28PAIVEE342 pKa = 4.25TLRR345 pKa = 11.84GIEE348 pKa = 4.14DD349 pKa = 3.19AHH351 pKa = 6.96GEE353 pKa = 4.18RR354 pKa = 11.84GSLPASDD361 pKa = 5.79AEE363 pKa = 4.35MKK365 pKa = 10.9NKK367 pKa = 9.93ILTSMVDD374 pKa = 3.37SIVNIKK380 pKa = 10.62KK381 pKa = 10.51EE382 pKa = 3.99GDD384 pKa = 3.48DD385 pKa = 4.65LYY387 pKa = 11.17IMLQGNGGYY396 pKa = 10.31DD397 pKa = 3.72KK398 pKa = 11.33QPVDD402 pKa = 3.49TDD404 pKa = 3.67QVEE407 pKa = 4.54SALSAMQSSSSDD419 pKa = 3.46LSSLIANMHH428 pKa = 6.46NGDD431 pKa = 3.46QYY433 pKa = 11.91YY434 pKa = 11.1LLGNTPVKK442 pKa = 10.64LGGPSTDD449 pKa = 2.94GATNITLGDD458 pKa = 3.49NDD460 pKa = 4.93SNVMFAGTTQDD471 pKa = 2.58WVTDD475 pKa = 4.22KK476 pKa = 11.35NGHH479 pKa = 5.0TSALEE484 pKa = 3.88DD485 pKa = 3.66HH486 pKa = 7.52DD487 pKa = 4.02GTFYY491 pKa = 10.7PSQHH495 pKa = 5.43MTSTMFGMGGDD506 pKa = 4.05DD507 pKa = 3.79IIYY510 pKa = 10.33GSFGEE515 pKa = 4.31DD516 pKa = 2.51CVYY519 pKa = 11.1AGDD522 pKa = 4.24GNDD525 pKa = 4.16MISTGAGNDD534 pKa = 3.98YY535 pKa = 10.98VWGDD539 pKa = 3.25AGDD542 pKa = 4.91DD543 pKa = 4.14YY544 pKa = 11.6ICGGRR549 pKa = 11.84GDD551 pKa = 3.94DD552 pKa = 3.93TLFGGDD558 pKa = 3.7GKK560 pKa = 10.75DD561 pKa = 2.8IFSFDD566 pKa = 3.83AEE568 pKa = 4.43SGHH571 pKa = 7.46DD572 pKa = 3.86IIRR575 pKa = 11.84DD576 pKa = 3.81FTPGEE581 pKa = 3.99DD582 pKa = 4.03KK583 pKa = 10.79IALVADD589 pKa = 3.32NWDD592 pKa = 3.79ALQSQIDD599 pKa = 3.79TSNANVPVITLNDD612 pKa = 3.65GSTVAVTLSNGQGLSEE628 pKa = 4.06NDD630 pKa = 3.86FYY632 pKa = 12.11YY633 pKa = 10.29MGANTALVFGHH644 pKa = 6.96TGTEE648 pKa = 4.38GNDD651 pKa = 3.58TIYY654 pKa = 11.28DD655 pKa = 3.71DD656 pKa = 3.92TSIVALRR663 pKa = 11.84GLSGDD668 pKa = 3.75DD669 pKa = 3.95TIHH672 pKa = 7.53CEE674 pKa = 3.51EE675 pKa = 5.49GYY677 pKa = 10.85NSGVFGDD684 pKa = 4.98DD685 pKa = 3.84GNDD688 pKa = 3.2TIYY691 pKa = 11.22ASNCSSGGFSGGAGDD706 pKa = 4.82DD707 pKa = 3.85RR708 pKa = 11.84IFAGSAGGVFFGDD721 pKa = 3.75AGDD724 pKa = 4.15DD725 pKa = 3.65SLYY728 pKa = 11.06GGSGEE733 pKa = 4.92DD734 pKa = 4.18DD735 pKa = 5.18FYY737 pKa = 11.5FQDD740 pKa = 3.95GFGHH744 pKa = 5.5DD745 pKa = 3.93TIYY748 pKa = 11.16GFAASTSMVANVYY761 pKa = 9.3EE762 pKa = 4.62PKK764 pKa = 10.75DD765 pKa = 3.67KK766 pKa = 10.71IALQGVSGLSDD777 pKa = 3.41WEE779 pKa = 4.15AVSNQITYY787 pKa = 10.18TDD789 pKa = 4.58LPDD792 pKa = 3.78SGYY795 pKa = 10.88CAIINDD801 pKa = 3.75GHH803 pKa = 7.17GNEE806 pKa = 4.32IDD808 pKa = 3.36IMLATGTQLVEE819 pKa = 3.93SDD821 pKa = 4.72FIFAA825 pKa = 5.1
MM1 pKa = 7.11LQRR4 pKa = 11.84RR5 pKa = 11.84EE6 pKa = 4.11LRR8 pKa = 11.84LLPPSLSISKK18 pKa = 10.72SMTGPHH24 pKa = 6.62LNLEE28 pKa = 4.25YY29 pKa = 10.35RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 11.01DD34 pKa = 4.17YY35 pKa = 11.33EE36 pKa = 4.24DD37 pKa = 3.6QPEE40 pKa = 4.43DD41 pKa = 3.72KK42 pKa = 10.98SIDD45 pKa = 3.17RR46 pKa = 11.84GTRR49 pKa = 11.84DD50 pKa = 3.2WIAAEE55 pKa = 4.01QPKK58 pKa = 10.11VYY60 pKa = 10.53HH61 pKa = 6.84FIFTVGDD68 pKa = 3.54DD69 pKa = 3.59RR70 pKa = 11.84LKK72 pKa = 10.34VTGTDD77 pKa = 3.45FDD79 pKa = 4.15TGFLADD85 pKa = 4.02MRR87 pKa = 11.84SGNDD91 pKa = 4.97FVTTWDD97 pKa = 3.72NPYY100 pKa = 10.8LPASTFWMGSGNDD113 pKa = 3.49VANVCGSSPTSDD125 pKa = 3.2EE126 pKa = 5.2DD127 pKa = 3.45EE128 pKa = 5.1DD129 pKa = 4.62EE130 pKa = 4.17EE131 pKa = 4.58VTRR134 pKa = 11.84VLCDD138 pKa = 4.51DD139 pKa = 4.95GDD141 pKa = 5.0DD142 pKa = 4.88GDD144 pKa = 5.75DD145 pKa = 4.37GDD147 pKa = 5.94DD148 pKa = 3.4IVNITGGGYY157 pKa = 11.0ANVFGDD163 pKa = 4.57DD164 pKa = 4.56GNDD167 pKa = 3.7TLCVQQNGAFKK178 pKa = 10.78SGVISLTGGAGYY190 pKa = 8.14DD191 pKa = 3.5TFLVTQHH198 pKa = 6.84PSSSYY203 pKa = 11.07NLPTSTGPASIGIFVTDD220 pKa = 3.12VLQDD224 pKa = 3.47VLGWVPGVGLATHH237 pKa = 7.18AAAALISTTTMAARR251 pKa = 11.84QSTRR255 pKa = 11.84FLNEE259 pKa = 3.15FSGAMDD265 pKa = 4.03GQDD268 pKa = 2.83GSVNIPQGGDD278 pKa = 3.17TYY280 pKa = 11.78VSIEE284 pKa = 4.47DD285 pKa = 3.92FDD287 pKa = 4.39PLEE290 pKa = 4.52DD291 pKa = 4.25QLVQTYY297 pKa = 8.29LHH299 pKa = 6.08NTNGMEE305 pKa = 4.06QYY307 pKa = 10.14SWQSQDD313 pKa = 3.17VASDD317 pKa = 3.38NFSIAFKK324 pKa = 10.98SDD326 pKa = 3.01GTNVSSVIHH335 pKa = 6.12VDD337 pKa = 3.28PAIVEE342 pKa = 4.25TLRR345 pKa = 11.84GIEE348 pKa = 4.14DD349 pKa = 3.19AHH351 pKa = 6.96GEE353 pKa = 4.18RR354 pKa = 11.84GSLPASDD361 pKa = 5.79AEE363 pKa = 4.35MKK365 pKa = 10.9NKK367 pKa = 9.93ILTSMVDD374 pKa = 3.37SIVNIKK380 pKa = 10.62KK381 pKa = 10.51EE382 pKa = 3.99GDD384 pKa = 3.48DD385 pKa = 4.65LYY387 pKa = 11.17IMLQGNGGYY396 pKa = 10.31DD397 pKa = 3.72KK398 pKa = 11.33QPVDD402 pKa = 3.49TDD404 pKa = 3.67QVEE407 pKa = 4.54SALSAMQSSSSDD419 pKa = 3.46LSSLIANMHH428 pKa = 6.46NGDD431 pKa = 3.46QYY433 pKa = 11.91YY434 pKa = 11.1LLGNTPVKK442 pKa = 10.64LGGPSTDD449 pKa = 2.94GATNITLGDD458 pKa = 3.49NDD460 pKa = 4.93SNVMFAGTTQDD471 pKa = 2.58WVTDD475 pKa = 4.22KK476 pKa = 11.35NGHH479 pKa = 5.0TSALEE484 pKa = 3.88DD485 pKa = 3.66HH486 pKa = 7.52DD487 pKa = 4.02GTFYY491 pKa = 10.7PSQHH495 pKa = 5.43MTSTMFGMGGDD506 pKa = 4.05DD507 pKa = 3.79IIYY510 pKa = 10.33GSFGEE515 pKa = 4.31DD516 pKa = 2.51CVYY519 pKa = 11.1AGDD522 pKa = 4.24GNDD525 pKa = 4.16MISTGAGNDD534 pKa = 3.98YY535 pKa = 10.98VWGDD539 pKa = 3.25AGDD542 pKa = 4.91DD543 pKa = 4.14YY544 pKa = 11.6ICGGRR549 pKa = 11.84GDD551 pKa = 3.94DD552 pKa = 3.93TLFGGDD558 pKa = 3.7GKK560 pKa = 10.75DD561 pKa = 2.8IFSFDD566 pKa = 3.83AEE568 pKa = 4.43SGHH571 pKa = 7.46DD572 pKa = 3.86IIRR575 pKa = 11.84DD576 pKa = 3.81FTPGEE581 pKa = 3.99DD582 pKa = 4.03KK583 pKa = 10.79IALVADD589 pKa = 3.32NWDD592 pKa = 3.79ALQSQIDD599 pKa = 3.79TSNANVPVITLNDD612 pKa = 3.65GSTVAVTLSNGQGLSEE628 pKa = 4.06NDD630 pKa = 3.86FYY632 pKa = 12.11YY633 pKa = 10.29MGANTALVFGHH644 pKa = 6.96TGTEE648 pKa = 4.38GNDD651 pKa = 3.58TIYY654 pKa = 11.28DD655 pKa = 3.71DD656 pKa = 3.92TSIVALRR663 pKa = 11.84GLSGDD668 pKa = 3.75DD669 pKa = 3.95TIHH672 pKa = 7.53CEE674 pKa = 3.51EE675 pKa = 5.49GYY677 pKa = 10.85NSGVFGDD684 pKa = 4.98DD685 pKa = 3.84GNDD688 pKa = 3.2TIYY691 pKa = 11.22ASNCSSGGFSGGAGDD706 pKa = 4.82DD707 pKa = 3.85RR708 pKa = 11.84IFAGSAGGVFFGDD721 pKa = 3.75AGDD724 pKa = 4.15DD725 pKa = 3.65SLYY728 pKa = 11.06GGSGEE733 pKa = 4.92DD734 pKa = 4.18DD735 pKa = 5.18FYY737 pKa = 11.5FQDD740 pKa = 3.95GFGHH744 pKa = 5.5DD745 pKa = 3.93TIYY748 pKa = 11.16GFAASTSMVANVYY761 pKa = 9.3EE762 pKa = 4.62PKK764 pKa = 10.75DD765 pKa = 3.67KK766 pKa = 10.71IALQGVSGLSDD777 pKa = 3.41WEE779 pKa = 4.15AVSNQITYY787 pKa = 10.18TDD789 pKa = 4.58LPDD792 pKa = 3.78SGYY795 pKa = 10.88CAIINDD801 pKa = 3.75GHH803 pKa = 7.17GNEE806 pKa = 4.32IDD808 pKa = 3.36IMLATGTQLVEE819 pKa = 3.93SDD821 pKa = 4.72FIFAA825 pKa = 5.1
Molecular weight: 87.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A506U755|A0A506U755_9RHIZ Dephospho-CoA kinase OS=Martelella sp. BGMRC2036 OX=2590451 GN=coaE PE=3 SV=1
MM1 pKa = 6.51STKK4 pKa = 9.4RR5 pKa = 11.84TYY7 pKa = 10.43QPSKK11 pKa = 9.73LVRR14 pKa = 11.84KK15 pKa = 9.15RR16 pKa = 11.84RR17 pKa = 11.84HH18 pKa = 4.42GFRR21 pKa = 11.84ARR23 pKa = 11.84MATKK27 pKa = 10.27GGRR30 pKa = 11.84AVLAARR36 pKa = 11.84RR37 pKa = 11.84TRR39 pKa = 11.84GRR41 pKa = 11.84KK42 pKa = 9.13RR43 pKa = 11.84LSAA46 pKa = 4.03
MM1 pKa = 6.51STKK4 pKa = 9.4RR5 pKa = 11.84TYY7 pKa = 10.43QPSKK11 pKa = 9.73LVRR14 pKa = 11.84KK15 pKa = 9.15RR16 pKa = 11.84RR17 pKa = 11.84HH18 pKa = 4.42GFRR21 pKa = 11.84ARR23 pKa = 11.84MATKK27 pKa = 10.27GGRR30 pKa = 11.84AVLAARR36 pKa = 11.84RR37 pKa = 11.84TRR39 pKa = 11.84GRR41 pKa = 11.84KK42 pKa = 9.13RR43 pKa = 11.84LSAA46 pKa = 4.03
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1450704 |
24 |
4640 |
330.2 |
35.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.964 ± 0.045 | 0.825 ± 0.011 |
5.942 ± 0.035 | 5.609 ± 0.034 |
4.064 ± 0.025 | 8.496 ± 0.068 |
1.974 ± 0.019 | 5.832 ± 0.025 |
3.549 ± 0.027 | 9.997 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.739 ± 0.019 | 2.906 ± 0.022 |
4.822 ± 0.031 | 3.01 ± 0.021 |
6.304 ± 0.043 | 5.842 ± 0.041 |
5.449 ± 0.034 | 7.058 ± 0.034 |
1.222 ± 0.017 | 2.397 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
