
Gemmata obscuriglobus
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Gemmatales; Gemmataceae; Gemmata
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
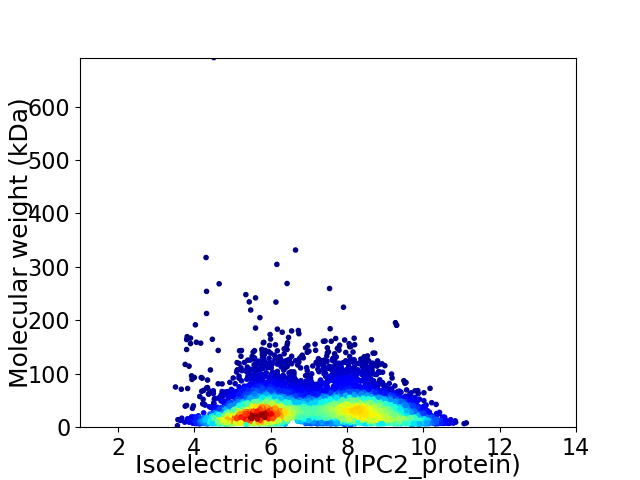
Virtual 2D-PAGE plot for 6809 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z3GZA3|A0A2Z3GZA3_9BACT FliMN_C domain-containing protein OS=Gemmata obscuriglobus OX=114 GN=C1280_10415 PE=3 SV=1
MM1 pKa = 7.38SSARR5 pKa = 11.84SFSHH9 pKa = 7.21RR10 pKa = 11.84APQFVPAVEE19 pKa = 4.09GLEE22 pKa = 5.01DD23 pKa = 3.51RR24 pKa = 11.84TTPAGNVTAYY34 pKa = 10.81VFDD37 pKa = 3.64KK38 pKa = 10.88VLYY41 pKa = 8.9VTGTNDD47 pKa = 3.71TDD49 pKa = 4.01QIWIAGAGEE58 pKa = 3.94NAVVIRR64 pKa = 11.84SLDD67 pKa = 3.31GATTINGQSSVYY79 pKa = 10.05IGGVKK84 pKa = 9.6QGYY87 pKa = 8.86HH88 pKa = 6.12INMGGGDD95 pKa = 4.15DD96 pKa = 3.88TLLVTGTRR104 pKa = 11.84SGGGLNVDD112 pKa = 3.51MGVGNDD118 pKa = 3.35VLGISDD124 pKa = 4.46AGHH127 pKa = 6.72KK128 pKa = 10.2GGSVLTGGAGNDD140 pKa = 3.39VFVLHH145 pKa = 7.23ASGFRR150 pKa = 11.84NGVSLNTGDD159 pKa = 5.34GDD161 pKa = 4.15DD162 pKa = 3.64QVLVSGVSAPAFALVNPSGTDD183 pKa = 3.32YY184 pKa = 11.33FDD186 pKa = 4.66APGSALGRR194 pKa = 11.84PSVVGGFVPGSPTATVPPTAAAADD218 pKa = 4.26TVAPTATVSTFSASVTSTAAVPFTVTFDD246 pKa = 3.53EE247 pKa = 4.87DD248 pKa = 3.98VTGFTAAGLTVSNGTVTAFGAVNARR273 pKa = 11.84TYY275 pKa = 9.24TFSVVPSGQGAVSVSVAAGAAQDD298 pKa = 3.61AAGNANTASNTVAVTFDD315 pKa = 3.43SVAPAVTVNSLTSSSATPTLTGTVDD340 pKa = 3.61DD341 pKa = 5.01PNATVSVTVNGQTYY355 pKa = 9.41KK356 pKa = 10.33ATVSSTTWSVTLTNPLTDD374 pKa = 3.37GTFTISVSATDD385 pKa = 3.17AAGNVGGTSLTDD397 pKa = 3.45GLVVSLAGLTVTVDD411 pKa = 3.47TLATNSTTPTITGTVSDD428 pKa = 3.96ATATVQVTVDD438 pKa = 3.39GQTYY442 pKa = 7.14TATVSGTTWSVAVTTALVEE461 pKa = 4.06GTYY464 pKa = 9.51TVSATATDD472 pKa = 3.76TNGNVATASPADD484 pKa = 3.73GLVIDD489 pKa = 4.96LTTPTVTANALTTNSATPTLTGTVDD514 pKa = 3.57DD515 pKa = 4.26STAAVQVTVDD525 pKa = 3.67GQAYY529 pKa = 6.05TATVSGTTWSVSVPTALIEE548 pKa = 4.03GTYY551 pKa = 8.93TISVTAMDD559 pKa = 3.75AAGNVGAASLTGGLVVDD576 pKa = 4.28LTVPTVTINALITNSTTPTLTGTVSDD602 pKa = 3.78STATVQVTVDD612 pKa = 3.38GQTYY616 pKa = 8.07PATVSGNTWTVAITTALVEE635 pKa = 4.34GAYY638 pKa = 8.15TASVTATDD646 pKa = 3.39VAGNVGTASLTDD658 pKa = 3.7GVVVDD663 pKa = 4.14LTVPTVAVDD672 pKa = 3.79TVPTVAGAVTGTAGDD687 pKa = 3.79TLAGVTGVAVTIYY700 pKa = 10.93DD701 pKa = 3.92SVAQAYY707 pKa = 10.3LNASRR712 pKa = 11.84TAFDD716 pKa = 3.33SATEE720 pKa = 3.9VWITAEE726 pKa = 3.99TADD729 pKa = 5.58AFATWSVTVPVAGSYY744 pKa = 9.4VVVAEE749 pKa = 4.31ATDD752 pKa = 4.06GAGNTATGNTTAVTSS767 pKa = 3.87
MM1 pKa = 7.38SSARR5 pKa = 11.84SFSHH9 pKa = 7.21RR10 pKa = 11.84APQFVPAVEE19 pKa = 4.09GLEE22 pKa = 5.01DD23 pKa = 3.51RR24 pKa = 11.84TTPAGNVTAYY34 pKa = 10.81VFDD37 pKa = 3.64KK38 pKa = 10.88VLYY41 pKa = 8.9VTGTNDD47 pKa = 3.71TDD49 pKa = 4.01QIWIAGAGEE58 pKa = 3.94NAVVIRR64 pKa = 11.84SLDD67 pKa = 3.31GATTINGQSSVYY79 pKa = 10.05IGGVKK84 pKa = 9.6QGYY87 pKa = 8.86HH88 pKa = 6.12INMGGGDD95 pKa = 4.15DD96 pKa = 3.88TLLVTGTRR104 pKa = 11.84SGGGLNVDD112 pKa = 3.51MGVGNDD118 pKa = 3.35VLGISDD124 pKa = 4.46AGHH127 pKa = 6.72KK128 pKa = 10.2GGSVLTGGAGNDD140 pKa = 3.39VFVLHH145 pKa = 7.23ASGFRR150 pKa = 11.84NGVSLNTGDD159 pKa = 5.34GDD161 pKa = 4.15DD162 pKa = 3.64QVLVSGVSAPAFALVNPSGTDD183 pKa = 3.32YY184 pKa = 11.33FDD186 pKa = 4.66APGSALGRR194 pKa = 11.84PSVVGGFVPGSPTATVPPTAAAADD218 pKa = 4.26TVAPTATVSTFSASVTSTAAVPFTVTFDD246 pKa = 3.53EE247 pKa = 4.87DD248 pKa = 3.98VTGFTAAGLTVSNGTVTAFGAVNARR273 pKa = 11.84TYY275 pKa = 9.24TFSVVPSGQGAVSVSVAAGAAQDD298 pKa = 3.61AAGNANTASNTVAVTFDD315 pKa = 3.43SVAPAVTVNSLTSSSATPTLTGTVDD340 pKa = 3.61DD341 pKa = 5.01PNATVSVTVNGQTYY355 pKa = 9.41KK356 pKa = 10.33ATVSSTTWSVTLTNPLTDD374 pKa = 3.37GTFTISVSATDD385 pKa = 3.17AAGNVGGTSLTDD397 pKa = 3.45GLVVSLAGLTVTVDD411 pKa = 3.47TLATNSTTPTITGTVSDD428 pKa = 3.96ATATVQVTVDD438 pKa = 3.39GQTYY442 pKa = 7.14TATVSGTTWSVAVTTALVEE461 pKa = 4.06GTYY464 pKa = 9.51TVSATATDD472 pKa = 3.76TNGNVATASPADD484 pKa = 3.73GLVIDD489 pKa = 4.96LTTPTVTANALTTNSATPTLTGTVDD514 pKa = 3.57DD515 pKa = 4.26STAAVQVTVDD525 pKa = 3.67GQAYY529 pKa = 6.05TATVSGTTWSVSVPTALIEE548 pKa = 4.03GTYY551 pKa = 8.93TISVTAMDD559 pKa = 3.75AAGNVGAASLTGGLVVDD576 pKa = 4.28LTVPTVTINALITNSTTPTLTGTVSDD602 pKa = 3.78STATVQVTVDD612 pKa = 3.38GQTYY616 pKa = 8.07PATVSGNTWTVAITTALVEE635 pKa = 4.34GAYY638 pKa = 8.15TASVTATDD646 pKa = 3.39VAGNVGTASLTDD658 pKa = 3.7GVVVDD663 pKa = 4.14LTVPTVAVDD672 pKa = 3.79TVPTVAGAVTGTAGDD687 pKa = 3.79TLAGVTGVAVTIYY700 pKa = 10.93DD701 pKa = 3.92SVAQAYY707 pKa = 10.3LNASRR712 pKa = 11.84TAFDD716 pKa = 3.33SATEE720 pKa = 3.9VWITAEE726 pKa = 3.99TADD729 pKa = 5.58AFATWSVTVPVAGSYY744 pKa = 9.4VVVAEE749 pKa = 4.31ATDD752 pKa = 4.06GAGNTATGNTTAVTSS767 pKa = 3.87
Molecular weight: 75.22 kDa
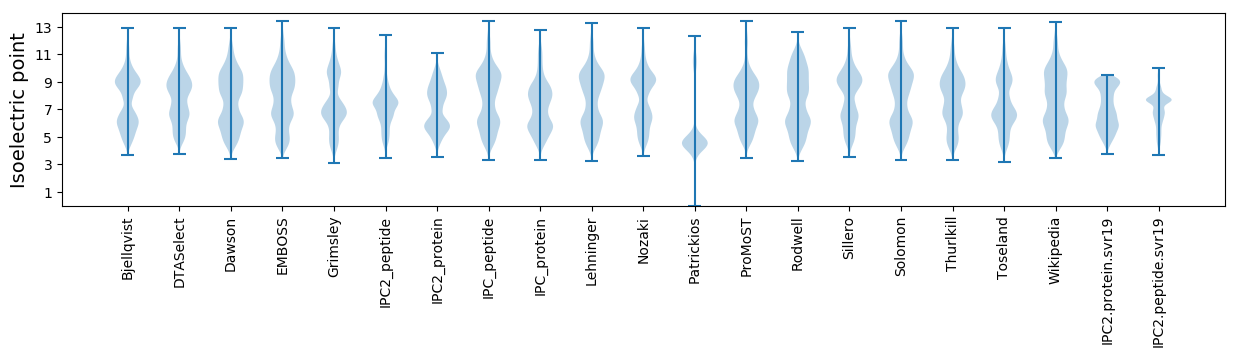
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z3GUD3|A0A2Z3GUD3_9BACT SBP_bac_5 domain-containing protein OS=Gemmata obscuriglobus OX=114 GN=C1280_14195 PE=4 SV=1
MM1 pKa = 7.23VGRR4 pKa = 11.84GRR6 pKa = 11.84ARR8 pKa = 11.84VGTRR12 pKa = 11.84RR13 pKa = 11.84FVSVVLRR20 pKa = 11.84VAPVGGGRR28 pKa = 11.84GSSPRR33 pKa = 11.84VAVVGGLRR41 pKa = 11.84CVATRR46 pKa = 11.84RR47 pKa = 11.84IGLGVLRR54 pKa = 11.84PAAPVARR61 pKa = 11.84APRR64 pKa = 11.84SRR66 pKa = 11.84GAARR70 pKa = 11.84VARR73 pKa = 11.84NRR75 pKa = 11.84GGQQ78 pKa = 3.06
MM1 pKa = 7.23VGRR4 pKa = 11.84GRR6 pKa = 11.84ARR8 pKa = 11.84VGTRR12 pKa = 11.84RR13 pKa = 11.84FVSVVLRR20 pKa = 11.84VAPVGGGRR28 pKa = 11.84GSSPRR33 pKa = 11.84VAVVGGLRR41 pKa = 11.84CVATRR46 pKa = 11.84RR47 pKa = 11.84IGLGVLRR54 pKa = 11.84PAAPVARR61 pKa = 11.84APRR64 pKa = 11.84SRR66 pKa = 11.84GAARR70 pKa = 11.84VARR73 pKa = 11.84NRR75 pKa = 11.84GGQQ78 pKa = 3.06
Molecular weight: 8.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2352850 |
26 |
6552 |
345.6 |
37.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.595 ± 0.048 | 1.06 ± 0.012 |
5.651 ± 0.019 | 5.582 ± 0.027 |
3.541 ± 0.014 | 8.759 ± 0.032 |
2.05 ± 0.013 | 3.255 ± 0.021 |
3.989 ± 0.031 | 9.93 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.655 ± 0.01 | 2.556 ± 0.021 |
6.408 ± 0.028 | 2.906 ± 0.018 |
7.501 ± 0.033 | 4.707 ± 0.024 |
5.81 ± 0.036 | 8.298 ± 0.024 |
1.52 ± 0.012 | 2.227 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
