
Macrococcus lamae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae; Macrococcus
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
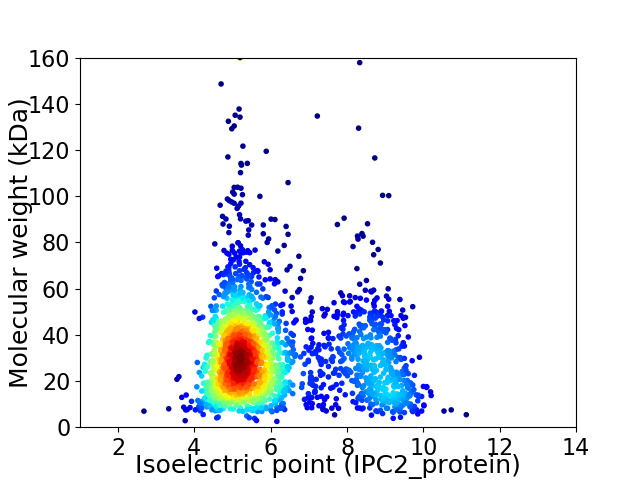
Virtual 2D-PAGE plot for 2076 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
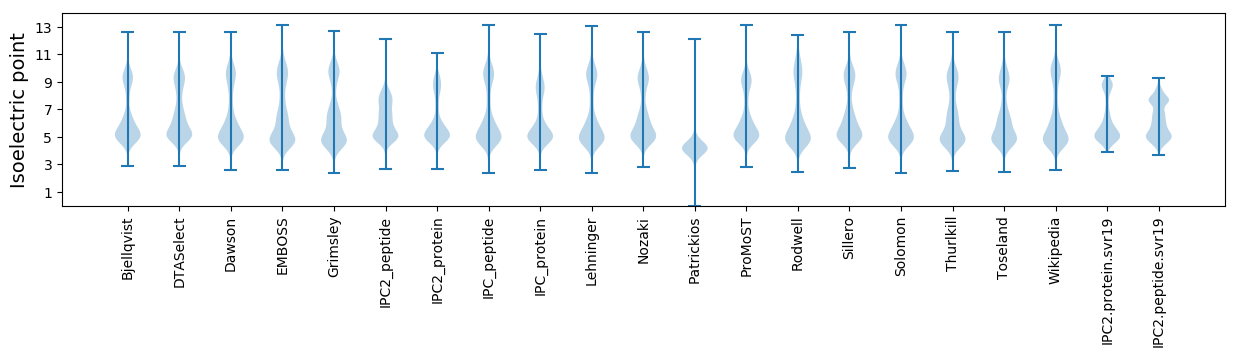
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6BXH3|A0A4R6BXH3_9STAP Tetratricopeptide repeat protein OS=Macrococcus lamae OX=198484 GN=ERX29_00230 PE=4 SV=1
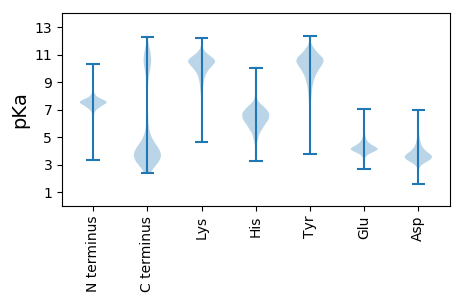
MM1 pKa = 8.2DD2 pKa = 5.25IYY4 pKa = 10.98HH5 pKa = 7.24IIDD8 pKa = 3.62KK9 pKa = 10.76VHH11 pKa = 5.88LQKK14 pKa = 10.23IEE16 pKa = 4.38QIDD19 pKa = 3.74NDD21 pKa = 3.81INDD24 pKa = 4.09ALMSEE29 pKa = 4.2DD30 pKa = 3.85HH31 pKa = 6.86EE32 pKa = 4.84GLFVLGEE39 pKa = 3.92TLYY42 pKa = 10.88QYY44 pKa = 11.8GLVPQGMKK52 pKa = 10.33VFEE55 pKa = 4.54ALYY58 pKa = 9.23HH59 pKa = 5.35QYY61 pKa = 10.58PEE63 pKa = 4.41EE64 pKa = 5.21IEE66 pKa = 4.12LLVYY70 pKa = 10.48YY71 pKa = 10.11IEE73 pKa = 4.6GLIDD77 pKa = 3.34EE78 pKa = 4.5NQIDD82 pKa = 3.81KK83 pKa = 11.05ALEE86 pKa = 4.17ILHH89 pKa = 6.5HH90 pKa = 6.32APKK93 pKa = 8.32TTEE96 pKa = 3.92RR97 pKa = 11.84LLLEE101 pKa = 4.45ADD103 pKa = 4.33VYY105 pKa = 10.15QQQDD109 pKa = 4.0MIEE112 pKa = 4.12VALDD116 pKa = 3.43KK117 pKa = 11.14LKK119 pKa = 10.92SAEE122 pKa = 4.09EE123 pKa = 4.03LAPDD127 pKa = 4.11DD128 pKa = 5.59LIITFALAEE137 pKa = 4.35LFYY140 pKa = 11.06FDD142 pKa = 5.07AQYY145 pKa = 11.06LQAARR150 pKa = 11.84RR151 pKa = 11.84YY152 pKa = 7.21EE153 pKa = 3.9TLLNSGEE160 pKa = 4.02QVINHH165 pKa = 5.4VNVNSRR171 pKa = 11.84LADD174 pKa = 3.43AMLQSGDD181 pKa = 3.67YY182 pKa = 8.96EE183 pKa = 3.91QAEE186 pKa = 4.58IYY188 pKa = 10.71FNTLEE193 pKa = 4.15EE194 pKa = 4.25QEE196 pKa = 4.25MTSDD200 pKa = 3.49DD201 pKa = 4.28FFRR204 pKa = 11.84KK205 pKa = 10.26AIALQKK211 pKa = 10.28NDD213 pKa = 4.33HH214 pKa = 6.43VPDD217 pKa = 5.7AIQVLNTLLEE227 pKa = 4.2KK228 pKa = 11.06DD229 pKa = 3.91PDD231 pKa = 3.84YY232 pKa = 11.06MHH234 pKa = 7.79AYY236 pKa = 9.82LLLVNLYY243 pKa = 9.16EE244 pKa = 4.62AEE246 pKa = 4.07RR247 pKa = 11.84LYY249 pKa = 11.32DD250 pKa = 3.65EE251 pKa = 6.14AILIGKK257 pKa = 8.97QGLALNEE264 pKa = 4.07FFKK267 pKa = 11.14EE268 pKa = 3.98LLVDD272 pKa = 3.82TARR275 pKa = 11.84VMLHH279 pKa = 6.07TKK281 pKa = 9.05NQEE284 pKa = 3.6AEE286 pKa = 4.01QYY288 pKa = 10.37LIQALTIDD296 pKa = 3.85PSYY299 pKa = 11.35TEE301 pKa = 4.1AALLLSDD308 pKa = 4.22YY309 pKa = 11.07YY310 pKa = 11.22KK311 pKa = 10.91EE312 pKa = 3.9NDD314 pKa = 3.33MFEE317 pKa = 4.01EE318 pKa = 4.42SVRR321 pKa = 11.84LFSLIDD327 pKa = 3.7EE328 pKa = 4.62EE329 pKa = 6.78DD330 pKa = 3.12IDD332 pKa = 6.51PIVKK336 pKa = 9.53WNLGYY341 pKa = 10.44SYY343 pKa = 10.89QQLEE347 pKa = 4.08RR348 pKa = 11.84DD349 pKa = 3.49NEE351 pKa = 4.06AKK353 pKa = 10.41HH354 pKa = 6.42FYY356 pKa = 8.7EE357 pKa = 4.4QAYY360 pKa = 10.68ADD362 pKa = 4.42LSDD365 pKa = 4.77NLDD368 pKa = 3.45FLYY371 pKa = 10.44DD372 pKa = 3.49YY373 pKa = 11.02HH374 pKa = 8.32DD375 pKa = 4.11YY376 pKa = 11.12LLEE379 pKa = 5.96IGEE382 pKa = 4.61DD383 pKa = 3.54TTKK386 pKa = 10.3VRR388 pKa = 11.84SQLTALDD395 pKa = 4.35PGFDD399 pKa = 3.42PYY401 pKa = 11.38EE402 pKa = 3.92RR403 pKa = 5.25
MM1 pKa = 8.2DD2 pKa = 5.25IYY4 pKa = 10.98HH5 pKa = 7.24IIDD8 pKa = 3.62KK9 pKa = 10.76VHH11 pKa = 5.88LQKK14 pKa = 10.23IEE16 pKa = 4.38QIDD19 pKa = 3.74NDD21 pKa = 3.81INDD24 pKa = 4.09ALMSEE29 pKa = 4.2DD30 pKa = 3.85HH31 pKa = 6.86EE32 pKa = 4.84GLFVLGEE39 pKa = 3.92TLYY42 pKa = 10.88QYY44 pKa = 11.8GLVPQGMKK52 pKa = 10.33VFEE55 pKa = 4.54ALYY58 pKa = 9.23HH59 pKa = 5.35QYY61 pKa = 10.58PEE63 pKa = 4.41EE64 pKa = 5.21IEE66 pKa = 4.12LLVYY70 pKa = 10.48YY71 pKa = 10.11IEE73 pKa = 4.6GLIDD77 pKa = 3.34EE78 pKa = 4.5NQIDD82 pKa = 3.81KK83 pKa = 11.05ALEE86 pKa = 4.17ILHH89 pKa = 6.5HH90 pKa = 6.32APKK93 pKa = 8.32TTEE96 pKa = 3.92RR97 pKa = 11.84LLLEE101 pKa = 4.45ADD103 pKa = 4.33VYY105 pKa = 10.15QQQDD109 pKa = 4.0MIEE112 pKa = 4.12VALDD116 pKa = 3.43KK117 pKa = 11.14LKK119 pKa = 10.92SAEE122 pKa = 4.09EE123 pKa = 4.03LAPDD127 pKa = 4.11DD128 pKa = 5.59LIITFALAEE137 pKa = 4.35LFYY140 pKa = 11.06FDD142 pKa = 5.07AQYY145 pKa = 11.06LQAARR150 pKa = 11.84RR151 pKa = 11.84YY152 pKa = 7.21EE153 pKa = 3.9TLLNSGEE160 pKa = 4.02QVINHH165 pKa = 5.4VNVNSRR171 pKa = 11.84LADD174 pKa = 3.43AMLQSGDD181 pKa = 3.67YY182 pKa = 8.96EE183 pKa = 3.91QAEE186 pKa = 4.58IYY188 pKa = 10.71FNTLEE193 pKa = 4.15EE194 pKa = 4.25QEE196 pKa = 4.25MTSDD200 pKa = 3.49DD201 pKa = 4.28FFRR204 pKa = 11.84KK205 pKa = 10.26AIALQKK211 pKa = 10.28NDD213 pKa = 4.33HH214 pKa = 6.43VPDD217 pKa = 5.7AIQVLNTLLEE227 pKa = 4.2KK228 pKa = 11.06DD229 pKa = 3.91PDD231 pKa = 3.84YY232 pKa = 11.06MHH234 pKa = 7.79AYY236 pKa = 9.82LLLVNLYY243 pKa = 9.16EE244 pKa = 4.62AEE246 pKa = 4.07RR247 pKa = 11.84LYY249 pKa = 11.32DD250 pKa = 3.65EE251 pKa = 6.14AILIGKK257 pKa = 8.97QGLALNEE264 pKa = 4.07FFKK267 pKa = 11.14EE268 pKa = 3.98LLVDD272 pKa = 3.82TARR275 pKa = 11.84VMLHH279 pKa = 6.07TKK281 pKa = 9.05NQEE284 pKa = 3.6AEE286 pKa = 4.01QYY288 pKa = 10.37LIQALTIDD296 pKa = 3.85PSYY299 pKa = 11.35TEE301 pKa = 4.1AALLLSDD308 pKa = 4.22YY309 pKa = 11.07YY310 pKa = 11.22KK311 pKa = 10.91EE312 pKa = 3.9NDD314 pKa = 3.33MFEE317 pKa = 4.01EE318 pKa = 4.42SVRR321 pKa = 11.84LFSLIDD327 pKa = 3.7EE328 pKa = 4.62EE329 pKa = 6.78DD330 pKa = 3.12IDD332 pKa = 6.51PIVKK336 pKa = 9.53WNLGYY341 pKa = 10.44SYY343 pKa = 10.89QQLEE347 pKa = 4.08RR348 pKa = 11.84DD349 pKa = 3.49NEE351 pKa = 4.06AKK353 pKa = 10.41HH354 pKa = 6.42FYY356 pKa = 8.7EE357 pKa = 4.4QAYY360 pKa = 10.68ADD362 pKa = 4.42LSDD365 pKa = 4.77NLDD368 pKa = 3.45FLYY371 pKa = 10.44DD372 pKa = 3.49YY373 pKa = 11.02HH374 pKa = 8.32DD375 pKa = 4.11YY376 pKa = 11.12LLEE379 pKa = 5.96IGEE382 pKa = 4.61DD383 pKa = 3.54TTKK386 pKa = 10.3VRR388 pKa = 11.84SQLTALDD395 pKa = 4.35PGFDD399 pKa = 3.42PYY401 pKa = 11.38EE402 pKa = 3.92RR403 pKa = 5.25
Molecular weight: 47.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6BTK1|A0A4R6BTK1_9STAP MFS transporter OS=Macrococcus lamae OX=198484 GN=ERX29_07815 PE=4 SV=1
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.66NGRR29 pKa = 11.84RR30 pKa = 11.84VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.49VKK3 pKa = 9.1RR4 pKa = 11.84TYY6 pKa = 10.35QPNKK10 pKa = 8.16RR11 pKa = 11.84KK12 pKa = 9.54HH13 pKa = 5.99SKK15 pKa = 8.91VHH17 pKa = 5.68GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 9.66NGRR29 pKa = 11.84RR30 pKa = 11.84VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
610545 |
21 |
1418 |
294.1 |
33.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.874 ± 0.054 | 0.644 ± 0.016 |
5.747 ± 0.045 | 7.027 ± 0.058 |
4.509 ± 0.049 | 6.378 ± 0.059 |
2.342 ± 0.03 | 8.137 ± 0.056 |
6.714 ± 0.053 | 9.573 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.037 ± 0.025 | 4.73 ± 0.036 |
3.368 ± 0.029 | 3.74 ± 0.032 |
4.055 ± 0.038 | 5.793 ± 0.038 |
5.723 ± 0.04 | 6.967 ± 0.043 |
0.781 ± 0.016 | 3.86 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
