
Lysobacter sp. Root96
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Lysobacter; unclassified Lysobacter
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
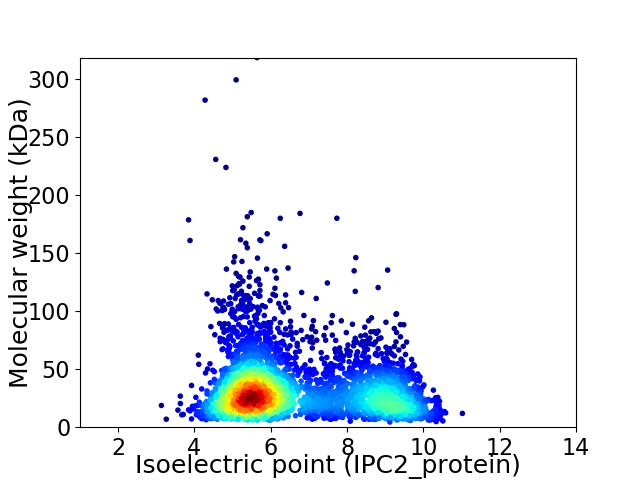
Virtual 2D-PAGE plot for 3827 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q9ERP3|A0A0Q9ERP3_9GAMM Phage_sheath_1 domain-containing protein OS=Lysobacter sp. Root96 OX=1736612 GN=ASE45_05395 PE=3 SV=1
MM1 pKa = 7.52AKK3 pKa = 10.1SVDD6 pKa = 3.48NANPVVGTNVVFTLTVTNNGPSASAGVSVADD37 pKa = 4.76LLPNGYY43 pKa = 7.98TYY45 pKa = 11.31VSDD48 pKa = 3.95NSAGAYY54 pKa = 9.87VPGTGVWTIGSLANAASTSLQITATVNATGTYY86 pKa = 10.29LNTATVSSTTNDD98 pKa = 3.2PTPANNTGTATTTPTASADD117 pKa = 3.59LSIDD121 pKa = 3.47KK122 pKa = 10.47DD123 pKa = 3.73DD124 pKa = 4.86AGASVVPGNNLVYY137 pKa = 10.14TIIVANAGPSAASNVVVSDD156 pKa = 5.14PIPAGLNFVSAAGAGWACVNNAGTVEE182 pKa = 4.25CSRR185 pKa = 11.84PTLAAGSNAAIALTLAVPANYY206 pKa = 10.18AGASSISNTASVGSATADD224 pKa = 3.45PNSGNDD230 pKa = 3.24SDD232 pKa = 5.19TEE234 pKa = 4.28TTPVSISIDD243 pKa = 3.64AVDD246 pKa = 4.53DD247 pKa = 4.08AATVLPFAGGTVPNVTVNDD266 pKa = 3.91TTNGVAVVIGVNATAPSIVSNGGLTGLIANPDD298 pKa = 3.19GSFTVPAGTAPGAYY312 pKa = 7.55TVGYY316 pKa = 9.07RR317 pKa = 11.84ICTLPATTPATCDD330 pKa = 3.5DD331 pKa = 4.16ANVPVVVGPNADD343 pKa = 4.49DD344 pKa = 5.13DD345 pKa = 5.4NEE347 pKa = 4.36TTPQNVALNGNASTNDD363 pKa = 3.34TASAGSVYY371 pKa = 10.93SLVSGPASGTLTFNADD387 pKa = 2.66GSYY390 pKa = 11.13VYY392 pKa = 10.25TPNTNFNGSDD402 pKa = 3.2SFNYY406 pKa = 9.18RR407 pKa = 11.84VCLPAPNGTVCDD419 pKa = 3.72TATVNITITAATIVAVDD436 pKa = 3.93DD437 pKa = 3.83TAATNFNTPVATNAVGNDD455 pKa = 3.49TTSGGVIDD463 pKa = 4.52PASVVATSGANGNVTCAAGSCTYY486 pKa = 10.81SPNPGFAGVDD496 pKa = 3.32SYY498 pKa = 12.07DD499 pKa = 3.55YY500 pKa = 9.36TVCLAAPNGTVCDD513 pKa = 3.67TATVNVVVGPNAGDD527 pKa = 3.82DD528 pKa = 4.75SEE530 pKa = 4.54TTPQNTALNATVTGNDD546 pKa = 3.46SAPAGAAYY554 pKa = 9.19TLVGGTANGTLTFNSDD570 pKa = 2.41GSYY573 pKa = 11.09VYY575 pKa = 10.01TPNANYY581 pKa = 10.44SGPDD585 pKa = 3.04SFNYY589 pKa = 9.64SVCLPAPNGTVCDD602 pKa = 3.87TATVTITVGGNTVTAVDD619 pKa = 5.01DD620 pKa = 4.5SASTPQNTPVGISVIGNDD638 pKa = 3.56TTTGAPLNPASVSVVTPPTSGSVSCDD664 pKa = 3.04SAGLCTYY671 pKa = 9.59TPTAGFSGNTSFVYY685 pKa = 9.83RR686 pKa = 11.84VCDD689 pKa = 3.38VSTPTPVCANATVTITVAQAVIVANDD715 pKa = 3.84DD716 pKa = 4.23DD717 pKa = 5.36YY718 pKa = 12.06SGSPVNGTNGSANVGNALTNDD739 pKa = 3.89SLNGGAIDD747 pKa = 4.06LAFVTLTVNDD757 pKa = 4.73PNPGDD762 pKa = 3.84GVAVASDD769 pKa = 3.65GTVSVAAGTPAGSYY783 pKa = 8.59VIPYY787 pKa = 7.42QLCEE791 pKa = 3.94TLHH794 pKa = 6.85PSNCDD799 pKa = 2.98PATITVVVNAAPIDD813 pKa = 4.0AVDD816 pKa = 4.84DD817 pKa = 4.24PVATPVNGSTGGTGVINVFGNDD839 pKa = 3.66TLNGAAATTTNVVLTPVSNGPLTVNANGTVDD870 pKa = 3.57VAPNTPAGVYY880 pKa = 7.57TVSYY884 pKa = 9.3RR885 pKa = 11.84ICEE888 pKa = 3.95VLNPSNCDD896 pKa = 3.13TALVTVTVNAAPIDD910 pKa = 3.93AVDD913 pKa = 4.58DD914 pKa = 4.27VVATPVNGSTGATAVVNVLGNDD936 pKa = 3.71TLNGSTVNPADD947 pKa = 3.56VTLTPVTSGPLTVNANGTVDD967 pKa = 3.57VAPNTPAGTYY977 pKa = 7.8TVTYY981 pKa = 8.18QICEE985 pKa = 3.89MLNPSNCDD993 pKa = 3.0TAVATVTVVAPAITAVDD1010 pKa = 3.88DD1011 pKa = 4.05PVGGGNGVTPQNTPLVTSVLGNDD1034 pKa = 4.11TLNGAPIDD1042 pKa = 3.69PTLINITIASNPANGTVTVNSNGTIVYY1069 pKa = 8.01TPNPNFSGNDD1079 pKa = 3.09SYY1081 pKa = 11.78TYY1083 pKa = 8.8TICEE1087 pKa = 4.15KK1088 pKa = 10.84LNPANCATATVSVLVQPNLVVATDD1112 pKa = 4.6DD1113 pKa = 3.59VTTTNQQVPVTISVLGNDD1131 pKa = 4.18TVTSAPLDD1139 pKa = 3.82PASVFVTVAPANGTVNCAAGVCTYY1163 pKa = 10.12TPNQFFAGTDD1173 pKa = 3.09SFTYY1177 pKa = 9.8RR1178 pKa = 11.84VCDD1181 pKa = 3.43TSVPTPVCDD1190 pKa = 3.17IAVVTITVAANAPVLRR1206 pKa = 11.84LSKK1209 pKa = 10.6QSAQRR1214 pKa = 11.84SVKK1217 pKa = 10.15IGDD1220 pKa = 3.57LVRR1223 pKa = 11.84YY1224 pKa = 6.96TVIAEE1229 pKa = 4.17NVGDD1233 pKa = 4.08SPARR1237 pKa = 11.84NATLVDD1243 pKa = 3.78TPPAGFTYY1251 pKa = 10.75VDD1253 pKa = 3.8GSLSVDD1259 pKa = 4.31DD1260 pKa = 5.24EE1261 pKa = 6.07DD1262 pKa = 4.94DD1263 pKa = 3.7AASIGGTNPLRR1274 pKa = 11.84INGIDD1279 pKa = 3.39VAIGQRR1285 pKa = 11.84ATIVYY1290 pKa = 8.18FLRR1293 pKa = 11.84VGAGVGKK1300 pKa = 10.03GVHH1303 pKa = 6.21SNSVTALDD1311 pKa = 4.44SNGSSIGNVATADD1324 pKa = 3.09VDD1326 pKa = 3.9MAGDD1330 pKa = 3.84PLLEE1334 pKa = 5.45DD1335 pKa = 3.79SLIMGSVYY1343 pKa = 10.66DD1344 pKa = 5.44DD1345 pKa = 3.81RR1346 pKa = 11.84DD1347 pKa = 3.32GDD1349 pKa = 3.82GWQDD1353 pKa = 3.2PAKK1356 pKa = 10.38AGGVRR1361 pKa = 11.84VQGGFAPGAYY1371 pKa = 9.25VAGSTTVDD1379 pKa = 3.5RR1380 pKa = 11.84GTGPQPEE1387 pKa = 4.96PDD1389 pKa = 3.18TSAPLLHH1396 pKa = 7.09GIAIGDD1402 pKa = 3.46IVGRR1406 pKa = 11.84SSSADD1411 pKa = 3.1DD1412 pKa = 3.82AQRR1415 pKa = 11.84HH1416 pKa = 4.29QVVVRR1421 pKa = 11.84QTLRR1425 pKa = 11.84SLDD1428 pKa = 3.77FSDD1431 pKa = 6.03DD1432 pKa = 3.67FVLTTDD1438 pKa = 3.43EE1439 pKa = 4.55GSSVRR1444 pKa = 11.84MNAAGQTTVDD1454 pKa = 3.71RR1455 pKa = 11.84VRR1457 pKa = 11.84GDD1459 pKa = 3.13AAKK1462 pKa = 10.82GLTAQDD1468 pKa = 3.7LQVTRR1473 pKa = 11.84QVSQVADD1480 pKa = 4.43GYY1482 pKa = 9.76EE1483 pKa = 3.68VSYY1486 pKa = 9.45TIRR1489 pKa = 11.84NEE1491 pKa = 4.49GIDD1494 pKa = 3.51EE1495 pKa = 4.26RR1496 pKa = 11.84GIPGVRR1502 pKa = 11.84IASVEE1507 pKa = 3.95GLIMQTDD1514 pKa = 3.99AYY1516 pKa = 10.35GRR1518 pKa = 11.84YY1519 pKa = 8.65HH1520 pKa = 7.97LEE1522 pKa = 4.71GISGGDD1528 pKa = 3.26AARR1531 pKa = 11.84GRR1533 pKa = 11.84NFILKK1538 pKa = 9.47VDD1540 pKa = 3.99QATLPPGSVFTTEE1553 pKa = 3.64NPRR1556 pKa = 11.84VRR1558 pKa = 11.84RR1559 pKa = 11.84VTQGVPTRR1567 pKa = 11.84FDD1569 pKa = 3.62FGVKK1573 pKa = 10.26LPAGEE1578 pKa = 3.93IRR1580 pKa = 11.84GGSSEE1585 pKa = 4.08TDD1587 pKa = 3.2VEE1589 pKa = 4.31LGEE1592 pKa = 4.39VMFEE1596 pKa = 4.05AGSDD1600 pKa = 3.8QVKK1603 pKa = 10.5SDD1605 pKa = 3.73YY1606 pKa = 11.59APMFGKK1612 pKa = 10.17IAQRR1616 pKa = 11.84LRR1618 pKa = 11.84DD1619 pKa = 3.64ADD1621 pKa = 3.92GGSVTIAAQAEE1632 pKa = 4.25AEE1634 pKa = 4.09ALAFARR1640 pKa = 11.84AHH1642 pKa = 6.13AVRR1645 pKa = 11.84AALSKK1650 pKa = 10.43EE1651 pKa = 4.11LEE1653 pKa = 4.09PALAAKK1659 pKa = 10.11VRR1661 pKa = 11.84IDD1663 pKa = 3.61VVAGTDD1669 pKa = 3.33ASALVSLDD1677 pKa = 3.31QAIKK1681 pKa = 10.6LGEE1684 pKa = 4.22LLFDD1688 pKa = 4.14TDD1690 pKa = 3.15QDD1692 pKa = 4.22TIRR1695 pKa = 11.84PQYY1698 pKa = 10.06RR1699 pKa = 11.84GLISEE1704 pKa = 4.17IAKK1707 pKa = 8.74TLNRR1711 pKa = 11.84EE1712 pKa = 3.63GRR1714 pKa = 11.84GVIGIVGRR1722 pKa = 11.84ADD1724 pKa = 3.21PRR1726 pKa = 11.84GANAYY1731 pKa = 9.23NVQLGLRR1738 pKa = 11.84RR1739 pKa = 11.84AKK1741 pKa = 10.56AVFEE1745 pKa = 4.85AISAEE1750 pKa = 4.12LEE1752 pKa = 4.14PKK1754 pKa = 10.0VKK1756 pKa = 10.48QKK1758 pKa = 10.35VRR1760 pKa = 11.84VDD1762 pKa = 3.04ITDD1765 pKa = 3.48DD1766 pKa = 3.31TNAPVGVGGRR1776 pKa = 3.6
MM1 pKa = 7.52AKK3 pKa = 10.1SVDD6 pKa = 3.48NANPVVGTNVVFTLTVTNNGPSASAGVSVADD37 pKa = 4.76LLPNGYY43 pKa = 7.98TYY45 pKa = 11.31VSDD48 pKa = 3.95NSAGAYY54 pKa = 9.87VPGTGVWTIGSLANAASTSLQITATVNATGTYY86 pKa = 10.29LNTATVSSTTNDD98 pKa = 3.2PTPANNTGTATTTPTASADD117 pKa = 3.59LSIDD121 pKa = 3.47KK122 pKa = 10.47DD123 pKa = 3.73DD124 pKa = 4.86AGASVVPGNNLVYY137 pKa = 10.14TIIVANAGPSAASNVVVSDD156 pKa = 5.14PIPAGLNFVSAAGAGWACVNNAGTVEE182 pKa = 4.25CSRR185 pKa = 11.84PTLAAGSNAAIALTLAVPANYY206 pKa = 10.18AGASSISNTASVGSATADD224 pKa = 3.45PNSGNDD230 pKa = 3.24SDD232 pKa = 5.19TEE234 pKa = 4.28TTPVSISIDD243 pKa = 3.64AVDD246 pKa = 4.53DD247 pKa = 4.08AATVLPFAGGTVPNVTVNDD266 pKa = 3.91TTNGVAVVIGVNATAPSIVSNGGLTGLIANPDD298 pKa = 3.19GSFTVPAGTAPGAYY312 pKa = 7.55TVGYY316 pKa = 9.07RR317 pKa = 11.84ICTLPATTPATCDD330 pKa = 3.5DD331 pKa = 4.16ANVPVVVGPNADD343 pKa = 4.49DD344 pKa = 5.13DD345 pKa = 5.4NEE347 pKa = 4.36TTPQNVALNGNASTNDD363 pKa = 3.34TASAGSVYY371 pKa = 10.93SLVSGPASGTLTFNADD387 pKa = 2.66GSYY390 pKa = 11.13VYY392 pKa = 10.25TPNTNFNGSDD402 pKa = 3.2SFNYY406 pKa = 9.18RR407 pKa = 11.84VCLPAPNGTVCDD419 pKa = 3.72TATVNITITAATIVAVDD436 pKa = 3.93DD437 pKa = 3.83TAATNFNTPVATNAVGNDD455 pKa = 3.49TTSGGVIDD463 pKa = 4.52PASVVATSGANGNVTCAAGSCTYY486 pKa = 10.81SPNPGFAGVDD496 pKa = 3.32SYY498 pKa = 12.07DD499 pKa = 3.55YY500 pKa = 9.36TVCLAAPNGTVCDD513 pKa = 3.67TATVNVVVGPNAGDD527 pKa = 3.82DD528 pKa = 4.75SEE530 pKa = 4.54TTPQNTALNATVTGNDD546 pKa = 3.46SAPAGAAYY554 pKa = 9.19TLVGGTANGTLTFNSDD570 pKa = 2.41GSYY573 pKa = 11.09VYY575 pKa = 10.01TPNANYY581 pKa = 10.44SGPDD585 pKa = 3.04SFNYY589 pKa = 9.64SVCLPAPNGTVCDD602 pKa = 3.87TATVTITVGGNTVTAVDD619 pKa = 5.01DD620 pKa = 4.5SASTPQNTPVGISVIGNDD638 pKa = 3.56TTTGAPLNPASVSVVTPPTSGSVSCDD664 pKa = 3.04SAGLCTYY671 pKa = 9.59TPTAGFSGNTSFVYY685 pKa = 9.83RR686 pKa = 11.84VCDD689 pKa = 3.38VSTPTPVCANATVTITVAQAVIVANDD715 pKa = 3.84DD716 pKa = 4.23DD717 pKa = 5.36YY718 pKa = 12.06SGSPVNGTNGSANVGNALTNDD739 pKa = 3.89SLNGGAIDD747 pKa = 4.06LAFVTLTVNDD757 pKa = 4.73PNPGDD762 pKa = 3.84GVAVASDD769 pKa = 3.65GTVSVAAGTPAGSYY783 pKa = 8.59VIPYY787 pKa = 7.42QLCEE791 pKa = 3.94TLHH794 pKa = 6.85PSNCDD799 pKa = 2.98PATITVVVNAAPIDD813 pKa = 4.0AVDD816 pKa = 4.84DD817 pKa = 4.24PVATPVNGSTGGTGVINVFGNDD839 pKa = 3.66TLNGAAATTTNVVLTPVSNGPLTVNANGTVDD870 pKa = 3.57VAPNTPAGVYY880 pKa = 7.57TVSYY884 pKa = 9.3RR885 pKa = 11.84ICEE888 pKa = 3.95VLNPSNCDD896 pKa = 3.13TALVTVTVNAAPIDD910 pKa = 3.93AVDD913 pKa = 4.58DD914 pKa = 4.27VVATPVNGSTGATAVVNVLGNDD936 pKa = 3.71TLNGSTVNPADD947 pKa = 3.56VTLTPVTSGPLTVNANGTVDD967 pKa = 3.57VAPNTPAGTYY977 pKa = 7.8TVTYY981 pKa = 8.18QICEE985 pKa = 3.89MLNPSNCDD993 pKa = 3.0TAVATVTVVAPAITAVDD1010 pKa = 3.88DD1011 pKa = 4.05PVGGGNGVTPQNTPLVTSVLGNDD1034 pKa = 4.11TLNGAPIDD1042 pKa = 3.69PTLINITIASNPANGTVTVNSNGTIVYY1069 pKa = 8.01TPNPNFSGNDD1079 pKa = 3.09SYY1081 pKa = 11.78TYY1083 pKa = 8.8TICEE1087 pKa = 4.15KK1088 pKa = 10.84LNPANCATATVSVLVQPNLVVATDD1112 pKa = 4.6DD1113 pKa = 3.59VTTTNQQVPVTISVLGNDD1131 pKa = 4.18TVTSAPLDD1139 pKa = 3.82PASVFVTVAPANGTVNCAAGVCTYY1163 pKa = 10.12TPNQFFAGTDD1173 pKa = 3.09SFTYY1177 pKa = 9.8RR1178 pKa = 11.84VCDD1181 pKa = 3.43TSVPTPVCDD1190 pKa = 3.17IAVVTITVAANAPVLRR1206 pKa = 11.84LSKK1209 pKa = 10.6QSAQRR1214 pKa = 11.84SVKK1217 pKa = 10.15IGDD1220 pKa = 3.57LVRR1223 pKa = 11.84YY1224 pKa = 6.96TVIAEE1229 pKa = 4.17NVGDD1233 pKa = 4.08SPARR1237 pKa = 11.84NATLVDD1243 pKa = 3.78TPPAGFTYY1251 pKa = 10.75VDD1253 pKa = 3.8GSLSVDD1259 pKa = 4.31DD1260 pKa = 5.24EE1261 pKa = 6.07DD1262 pKa = 4.94DD1263 pKa = 3.7AASIGGTNPLRR1274 pKa = 11.84INGIDD1279 pKa = 3.39VAIGQRR1285 pKa = 11.84ATIVYY1290 pKa = 8.18FLRR1293 pKa = 11.84VGAGVGKK1300 pKa = 10.03GVHH1303 pKa = 6.21SNSVTALDD1311 pKa = 4.44SNGSSIGNVATADD1324 pKa = 3.09VDD1326 pKa = 3.9MAGDD1330 pKa = 3.84PLLEE1334 pKa = 5.45DD1335 pKa = 3.79SLIMGSVYY1343 pKa = 10.66DD1344 pKa = 5.44DD1345 pKa = 3.81RR1346 pKa = 11.84DD1347 pKa = 3.32GDD1349 pKa = 3.82GWQDD1353 pKa = 3.2PAKK1356 pKa = 10.38AGGVRR1361 pKa = 11.84VQGGFAPGAYY1371 pKa = 9.25VAGSTTVDD1379 pKa = 3.5RR1380 pKa = 11.84GTGPQPEE1387 pKa = 4.96PDD1389 pKa = 3.18TSAPLLHH1396 pKa = 7.09GIAIGDD1402 pKa = 3.46IVGRR1406 pKa = 11.84SSSADD1411 pKa = 3.1DD1412 pKa = 3.82AQRR1415 pKa = 11.84HH1416 pKa = 4.29QVVVRR1421 pKa = 11.84QTLRR1425 pKa = 11.84SLDD1428 pKa = 3.77FSDD1431 pKa = 6.03DD1432 pKa = 3.67FVLTTDD1438 pKa = 3.43EE1439 pKa = 4.55GSSVRR1444 pKa = 11.84MNAAGQTTVDD1454 pKa = 3.71RR1455 pKa = 11.84VRR1457 pKa = 11.84GDD1459 pKa = 3.13AAKK1462 pKa = 10.82GLTAQDD1468 pKa = 3.7LQVTRR1473 pKa = 11.84QVSQVADD1480 pKa = 4.43GYY1482 pKa = 9.76EE1483 pKa = 3.68VSYY1486 pKa = 9.45TIRR1489 pKa = 11.84NEE1491 pKa = 4.49GIDD1494 pKa = 3.51EE1495 pKa = 4.26RR1496 pKa = 11.84GIPGVRR1502 pKa = 11.84IASVEE1507 pKa = 3.95GLIMQTDD1514 pKa = 3.99AYY1516 pKa = 10.35GRR1518 pKa = 11.84YY1519 pKa = 8.65HH1520 pKa = 7.97LEE1522 pKa = 4.71GISGGDD1528 pKa = 3.26AARR1531 pKa = 11.84GRR1533 pKa = 11.84NFILKK1538 pKa = 9.47VDD1540 pKa = 3.99QATLPPGSVFTTEE1553 pKa = 3.64NPRR1556 pKa = 11.84VRR1558 pKa = 11.84RR1559 pKa = 11.84VTQGVPTRR1567 pKa = 11.84FDD1569 pKa = 3.62FGVKK1573 pKa = 10.26LPAGEE1578 pKa = 3.93IRR1580 pKa = 11.84GGSSEE1585 pKa = 4.08TDD1587 pKa = 3.2VEE1589 pKa = 4.31LGEE1592 pKa = 4.39VMFEE1596 pKa = 4.05AGSDD1600 pKa = 3.8QVKK1603 pKa = 10.5SDD1605 pKa = 3.73YY1606 pKa = 11.59APMFGKK1612 pKa = 10.17IAQRR1616 pKa = 11.84LRR1618 pKa = 11.84DD1619 pKa = 3.64ADD1621 pKa = 3.92GGSVTIAAQAEE1632 pKa = 4.25AEE1634 pKa = 4.09ALAFARR1640 pKa = 11.84AHH1642 pKa = 6.13AVRR1645 pKa = 11.84AALSKK1650 pKa = 10.43EE1651 pKa = 4.11LEE1653 pKa = 4.09PALAAKK1659 pKa = 10.11VRR1661 pKa = 11.84IDD1663 pKa = 3.61VVAGTDD1669 pKa = 3.33ASALVSLDD1677 pKa = 3.31QAIKK1681 pKa = 10.6LGEE1684 pKa = 4.22LLFDD1688 pKa = 4.14TDD1690 pKa = 3.15QDD1692 pKa = 4.22TIRR1695 pKa = 11.84PQYY1698 pKa = 10.06RR1699 pKa = 11.84GLISEE1704 pKa = 4.17IAKK1707 pKa = 8.74TLNRR1711 pKa = 11.84EE1712 pKa = 3.63GRR1714 pKa = 11.84GVIGIVGRR1722 pKa = 11.84ADD1724 pKa = 3.21PRR1726 pKa = 11.84GANAYY1731 pKa = 9.23NVQLGLRR1738 pKa = 11.84RR1739 pKa = 11.84AKK1741 pKa = 10.56AVFEE1745 pKa = 4.85AISAEE1750 pKa = 4.12LEE1752 pKa = 4.14PKK1754 pKa = 10.0VKK1756 pKa = 10.48QKK1758 pKa = 10.35VRR1760 pKa = 11.84VDD1762 pKa = 3.04ITDD1765 pKa = 3.48DD1766 pKa = 3.31TNAPVGVGGRR1776 pKa = 3.6
Molecular weight: 178.54 kDa
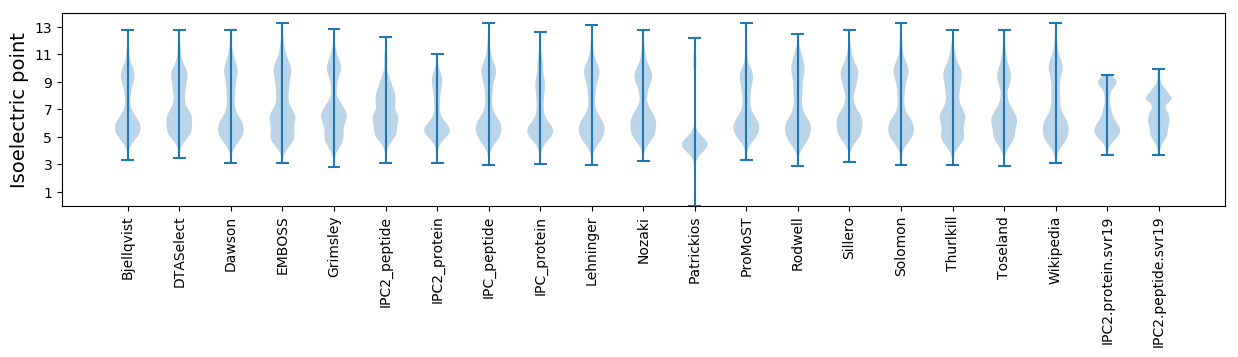
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q9EMA4|A0A0Q9EMA4_9GAMM Chaperone protein ClpB OS=Lysobacter sp. Root96 OX=1736612 GN=clpB PE=3 SV=1
MM1 pKa = 6.66STGRR5 pKa = 11.84RR6 pKa = 11.84LASRR10 pKa = 11.84AIAWQAGATALTALAFLPLGARR32 pKa = 11.84FALAAAVGGGAVFAGAFVAAWMALGGGVQPAGAAIGRR69 pKa = 11.84LLAGVMFKK77 pKa = 10.19WVVVFLVLGLGLAVFRR93 pKa = 11.84LPPLPMLVGVLAATLAFVLANLLKK117 pKa = 10.59RR118 pKa = 4.01
MM1 pKa = 6.66STGRR5 pKa = 11.84RR6 pKa = 11.84LASRR10 pKa = 11.84AIAWQAGATALTALAFLPLGARR32 pKa = 11.84FALAAAVGGGAVFAGAFVAAWMALGGGVQPAGAAIGRR69 pKa = 11.84LLAGVMFKK77 pKa = 10.19WVVVFLVLGLGLAVFRR93 pKa = 11.84LPPLPMLVGVLAATLAFVLANLLKK117 pKa = 10.59RR118 pKa = 4.01
Molecular weight: 11.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1228680 |
41 |
2854 |
321.1 |
34.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.55 ± 0.056 | 0.834 ± 0.012 |
5.931 ± 0.034 | 5.317 ± 0.038 |
3.291 ± 0.027 | 8.629 ± 0.038 |
2.11 ± 0.019 | 4.087 ± 0.028 |
2.72 ± 0.032 | 10.709 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.033 ± 0.019 | 2.514 ± 0.031 |
5.409 ± 0.029 | 3.641 ± 0.027 |
7.858 ± 0.041 | 5.269 ± 0.033 |
4.788 ± 0.042 | 7.219 ± 0.037 |
1.548 ± 0.02 | 2.541 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
