
Lake Sarah-associated circular virus-44
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.76
Get precalculated fractions of proteins
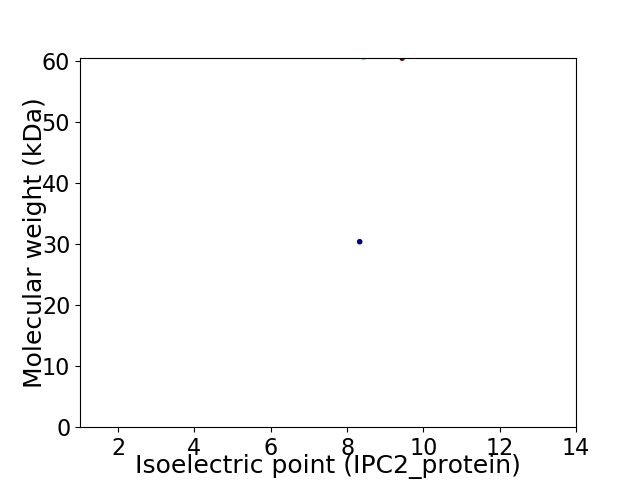
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
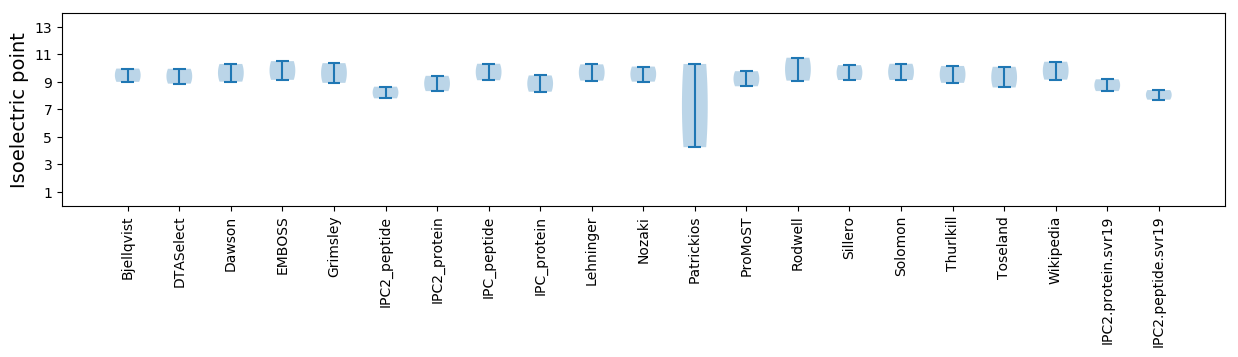
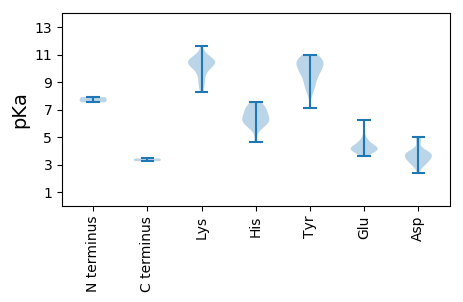
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQQ9|A0A140AQQ9_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-44 OX=1685773 PE=4 SV=1
MM1 pKa = 7.53EE2 pKa = 6.25KK3 pKa = 9.62IRR5 pKa = 11.84YY6 pKa = 8.25YY7 pKa = 10.36ILTIPHH13 pKa = 5.85YY14 pKa = 10.38AYY16 pKa = 9.73TPFLHH21 pKa = 6.5TSCNYY26 pKa = 9.59VRR28 pKa = 11.84GQLEE32 pKa = 4.04QGASGYY38 pKa = 8.25LHH40 pKa = 5.98WQMVVFLKK48 pKa = 10.69SPQRR52 pKa = 11.84RR53 pKa = 11.84SFVKK57 pKa = 10.29KK58 pKa = 10.36LYY60 pKa = 10.52GDD62 pKa = 3.69EE63 pKa = 4.09AHH65 pKa = 7.2IEE67 pKa = 4.16PTRR70 pKa = 11.84SAAAMEE76 pKa = 4.42YY77 pKa = 9.72VWKK80 pKa = 10.8QEE82 pKa = 3.77TSVPNTQFEE91 pKa = 4.78LGSLPTSRR99 pKa = 11.84CSSKK103 pKa = 10.83DD104 pKa = 2.42WDD106 pKa = 4.79AIRR109 pKa = 11.84SSAVEE114 pKa = 3.9GRR116 pKa = 11.84MEE118 pKa = 5.09SIPSDD123 pKa = 3.79VYY125 pKa = 10.08IRR127 pKa = 11.84CYY129 pKa = 11.0NSLKK133 pKa = 10.44RR134 pKa = 11.84IAVDD138 pKa = 3.07HH139 pKa = 6.4SKK141 pKa = 10.37PVCIEE146 pKa = 3.65RR147 pKa = 11.84KK148 pKa = 9.19IIVYY152 pKa = 8.85YY153 pKa = 10.86GPTGTGKK160 pKa = 9.96SRR162 pKa = 11.84RR163 pKa = 11.84AWDD166 pKa = 3.3EE167 pKa = 3.98AGFDD171 pKa = 4.8AYY173 pKa = 10.07PKK175 pKa = 10.72DD176 pKa = 3.98PNTKK180 pKa = 9.66FWDD183 pKa = 4.66GYY185 pKa = 9.71NGHH188 pKa = 6.78KK189 pKa = 10.52NVVIDD194 pKa = 3.86EE195 pKa = 4.11FRR197 pKa = 11.84GIINISNVLRR207 pKa = 11.84WFDD210 pKa = 3.77RR211 pKa = 11.84YY212 pKa = 9.66PVLVEE217 pKa = 4.1VKK219 pKa = 10.41GSSAVFKK226 pKa = 11.22AEE228 pKa = 4.29NIWITSNLHH237 pKa = 5.82PDD239 pKa = 2.94DD240 pKa = 4.98WYY242 pKa = 10.7PEE244 pKa = 4.25LDD246 pKa = 3.79PATKK250 pKa = 10.26LALKK254 pKa = 10.35RR255 pKa = 11.84RR256 pKa = 11.84LLIVNILL263 pKa = 3.44
MM1 pKa = 7.53EE2 pKa = 6.25KK3 pKa = 9.62IRR5 pKa = 11.84YY6 pKa = 8.25YY7 pKa = 10.36ILTIPHH13 pKa = 5.85YY14 pKa = 10.38AYY16 pKa = 9.73TPFLHH21 pKa = 6.5TSCNYY26 pKa = 9.59VRR28 pKa = 11.84GQLEE32 pKa = 4.04QGASGYY38 pKa = 8.25LHH40 pKa = 5.98WQMVVFLKK48 pKa = 10.69SPQRR52 pKa = 11.84RR53 pKa = 11.84SFVKK57 pKa = 10.29KK58 pKa = 10.36LYY60 pKa = 10.52GDD62 pKa = 3.69EE63 pKa = 4.09AHH65 pKa = 7.2IEE67 pKa = 4.16PTRR70 pKa = 11.84SAAAMEE76 pKa = 4.42YY77 pKa = 9.72VWKK80 pKa = 10.8QEE82 pKa = 3.77TSVPNTQFEE91 pKa = 4.78LGSLPTSRR99 pKa = 11.84CSSKK103 pKa = 10.83DD104 pKa = 2.42WDD106 pKa = 4.79AIRR109 pKa = 11.84SSAVEE114 pKa = 3.9GRR116 pKa = 11.84MEE118 pKa = 5.09SIPSDD123 pKa = 3.79VYY125 pKa = 10.08IRR127 pKa = 11.84CYY129 pKa = 11.0NSLKK133 pKa = 10.44RR134 pKa = 11.84IAVDD138 pKa = 3.07HH139 pKa = 6.4SKK141 pKa = 10.37PVCIEE146 pKa = 3.65RR147 pKa = 11.84KK148 pKa = 9.19IIVYY152 pKa = 8.85YY153 pKa = 10.86GPTGTGKK160 pKa = 9.96SRR162 pKa = 11.84RR163 pKa = 11.84AWDD166 pKa = 3.3EE167 pKa = 3.98AGFDD171 pKa = 4.8AYY173 pKa = 10.07PKK175 pKa = 10.72DD176 pKa = 3.98PNTKK180 pKa = 9.66FWDD183 pKa = 4.66GYY185 pKa = 9.71NGHH188 pKa = 6.78KK189 pKa = 10.52NVVIDD194 pKa = 3.86EE195 pKa = 4.11FRR197 pKa = 11.84GIINISNVLRR207 pKa = 11.84WFDD210 pKa = 3.77RR211 pKa = 11.84YY212 pKa = 9.66PVLVEE217 pKa = 4.1VKK219 pKa = 10.41GSSAVFKK226 pKa = 11.22AEE228 pKa = 4.29NIWITSNLHH237 pKa = 5.82PDD239 pKa = 2.94DD240 pKa = 4.98WYY242 pKa = 10.7PEE244 pKa = 4.25LDD246 pKa = 3.79PATKK250 pKa = 10.26LALKK254 pKa = 10.35RR255 pKa = 11.84RR256 pKa = 11.84LLIVNILL263 pKa = 3.44
Molecular weight: 30.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQQ9|A0A140AQQ9_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-44 OX=1685773 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84FGWWPHH8 pKa = 4.66HH9 pKa = 6.33HH10 pKa = 7.13CGPGNYY16 pKa = 8.63IKK18 pKa = 10.44SYY20 pKa = 10.76RR21 pKa = 11.84SLHH24 pKa = 6.17DD25 pKa = 3.26HH26 pKa = 7.03TDD28 pKa = 3.77NICRR32 pKa = 11.84DD33 pKa = 2.87HH34 pKa = 7.21DD35 pKa = 3.65KK36 pKa = 11.02AYY38 pKa = 10.72RR39 pKa = 11.84RR40 pKa = 11.84YY41 pKa = 9.88GNKK44 pKa = 10.14AYY46 pKa = 9.62IYY48 pKa = 9.95YY49 pKa = 10.59NKK51 pKa = 10.54ADD53 pKa = 3.5DD54 pKa = 3.81RR55 pKa = 11.84FIRR58 pKa = 11.84RR59 pKa = 11.84MDD61 pKa = 3.1RR62 pKa = 11.84RR63 pKa = 11.84KK64 pKa = 10.4GVWPKK69 pKa = 10.27IYY71 pKa = 10.53AGYY74 pKa = 10.34FKK76 pKa = 10.69GKK78 pKa = 10.01KK79 pKa = 8.87RR80 pKa = 11.84VAPVMDD86 pKa = 4.19EE87 pKa = 3.8PRR89 pKa = 11.84KK90 pKa = 9.48KK91 pKa = 10.34RR92 pKa = 11.84KK93 pKa = 10.14LEE95 pKa = 3.83VSFSSSKK102 pKa = 10.64NDD104 pKa = 3.54EE105 pKa = 4.09NSEE108 pKa = 4.2MPRR111 pKa = 11.84SRR113 pKa = 11.84SRR115 pKa = 11.84GRR117 pKa = 11.84TPTRR121 pKa = 11.84SKK123 pKa = 11.35SPMPTPRR130 pKa = 11.84KK131 pKa = 9.54SRR133 pKa = 11.84SPKK136 pKa = 8.88KK137 pKa = 8.37TYY139 pKa = 9.07QSRR142 pKa = 11.84GTSTDD147 pKa = 3.35SIRR150 pKa = 11.84NRR152 pKa = 11.84SMSSGRR158 pKa = 11.84ASSSKK163 pKa = 9.17ATAVIAASSTDD174 pKa = 3.35VVRR177 pKa = 11.84ARR179 pKa = 11.84TTKK182 pKa = 10.15KK183 pKa = 10.75GRR185 pKa = 11.84ILDD188 pKa = 3.78KK189 pKa = 10.52GTPNLLAGNLVAKK202 pKa = 9.44TDD204 pKa = 3.44KK205 pKa = 10.79KK206 pKa = 9.81FNKK209 pKa = 8.73MSYY212 pKa = 9.34PGWVHH217 pKa = 6.55EE218 pKa = 4.31NVTTGAAVTDD228 pKa = 3.9AQAIIISHH236 pKa = 6.26TDD238 pKa = 2.87MAQRR242 pKa = 11.84NVIQTVARR250 pKa = 11.84ALVKK254 pKa = 10.55SLWRR258 pKa = 11.84KK259 pKa = 8.31CGVYY263 pKa = 8.67VTSLEE268 pKa = 4.19QPIPPAWAVAPFPTARR284 pKa = 11.84LEE286 pKa = 4.09IRR288 pKa = 11.84DD289 pKa = 4.01LVSNGATVAVWTYY302 pKa = 9.1TATALSTFNDD312 pKa = 3.22WCVSLSTSIEE322 pKa = 4.22TYY324 pKa = 10.32LVSNLYY330 pKa = 10.55GSWAPAIMTCSPKK343 pKa = 10.65GNNGLTVEE351 pKa = 4.73FDD353 pKa = 3.32LSKK356 pKa = 11.64AEE358 pKa = 4.16VTVLTSNILKK368 pKa = 10.15VKK370 pKa = 10.15NISVDD375 pKa = 3.4GTNDD379 pKa = 3.67SILDD383 pKa = 3.63VNASPFIGKK392 pKa = 10.04GYY394 pKa = 10.15IIGGTNIEE402 pKa = 4.44FKK404 pKa = 10.52GYY406 pKa = 9.31PYY408 pKa = 10.85VIGGDD413 pKa = 3.94LNFSPDD419 pKa = 3.19TDD421 pKa = 2.87SGYY424 pKa = 8.62TVKK427 pKa = 9.85GTASVGGTKK436 pKa = 8.96IDD438 pKa = 3.8RR439 pKa = 11.84PPLKK443 pKa = 10.03ATIGRR448 pKa = 11.84CRR450 pKa = 11.84SMGNIGMNVGEE461 pKa = 4.69IKK463 pKa = 10.57SHH465 pKa = 5.75IAGYY469 pKa = 7.1SHH471 pKa = 7.56KK472 pKa = 10.89GNLWTFLTKK481 pKa = 10.65LGVRR485 pKa = 11.84SSTNAAPINLIQTRR499 pKa = 11.84SIGTTYY505 pKa = 10.18MFYY508 pKa = 10.57LYY510 pKa = 10.56KK511 pKa = 10.63QIFTDD516 pKa = 3.48NGNLSVEE523 pKa = 4.56AEE525 pKa = 4.28HH526 pKa = 6.99NMTTGVIVNLKK537 pKa = 8.58PTIHH541 pKa = 6.18SCKK544 pKa = 10.28SVNN547 pKa = 3.24
MM1 pKa = 7.9RR2 pKa = 11.84FGWWPHH8 pKa = 4.66HH9 pKa = 6.33HH10 pKa = 7.13CGPGNYY16 pKa = 8.63IKK18 pKa = 10.44SYY20 pKa = 10.76RR21 pKa = 11.84SLHH24 pKa = 6.17DD25 pKa = 3.26HH26 pKa = 7.03TDD28 pKa = 3.77NICRR32 pKa = 11.84DD33 pKa = 2.87HH34 pKa = 7.21DD35 pKa = 3.65KK36 pKa = 11.02AYY38 pKa = 10.72RR39 pKa = 11.84RR40 pKa = 11.84YY41 pKa = 9.88GNKK44 pKa = 10.14AYY46 pKa = 9.62IYY48 pKa = 9.95YY49 pKa = 10.59NKK51 pKa = 10.54ADD53 pKa = 3.5DD54 pKa = 3.81RR55 pKa = 11.84FIRR58 pKa = 11.84RR59 pKa = 11.84MDD61 pKa = 3.1RR62 pKa = 11.84RR63 pKa = 11.84KK64 pKa = 10.4GVWPKK69 pKa = 10.27IYY71 pKa = 10.53AGYY74 pKa = 10.34FKK76 pKa = 10.69GKK78 pKa = 10.01KK79 pKa = 8.87RR80 pKa = 11.84VAPVMDD86 pKa = 4.19EE87 pKa = 3.8PRR89 pKa = 11.84KK90 pKa = 9.48KK91 pKa = 10.34RR92 pKa = 11.84KK93 pKa = 10.14LEE95 pKa = 3.83VSFSSSKK102 pKa = 10.64NDD104 pKa = 3.54EE105 pKa = 4.09NSEE108 pKa = 4.2MPRR111 pKa = 11.84SRR113 pKa = 11.84SRR115 pKa = 11.84GRR117 pKa = 11.84TPTRR121 pKa = 11.84SKK123 pKa = 11.35SPMPTPRR130 pKa = 11.84KK131 pKa = 9.54SRR133 pKa = 11.84SPKK136 pKa = 8.88KK137 pKa = 8.37TYY139 pKa = 9.07QSRR142 pKa = 11.84GTSTDD147 pKa = 3.35SIRR150 pKa = 11.84NRR152 pKa = 11.84SMSSGRR158 pKa = 11.84ASSSKK163 pKa = 9.17ATAVIAASSTDD174 pKa = 3.35VVRR177 pKa = 11.84ARR179 pKa = 11.84TTKK182 pKa = 10.15KK183 pKa = 10.75GRR185 pKa = 11.84ILDD188 pKa = 3.78KK189 pKa = 10.52GTPNLLAGNLVAKK202 pKa = 9.44TDD204 pKa = 3.44KK205 pKa = 10.79KK206 pKa = 9.81FNKK209 pKa = 8.73MSYY212 pKa = 9.34PGWVHH217 pKa = 6.55EE218 pKa = 4.31NVTTGAAVTDD228 pKa = 3.9AQAIIISHH236 pKa = 6.26TDD238 pKa = 2.87MAQRR242 pKa = 11.84NVIQTVARR250 pKa = 11.84ALVKK254 pKa = 10.55SLWRR258 pKa = 11.84KK259 pKa = 8.31CGVYY263 pKa = 8.67VTSLEE268 pKa = 4.19QPIPPAWAVAPFPTARR284 pKa = 11.84LEE286 pKa = 4.09IRR288 pKa = 11.84DD289 pKa = 4.01LVSNGATVAVWTYY302 pKa = 9.1TATALSTFNDD312 pKa = 3.22WCVSLSTSIEE322 pKa = 4.22TYY324 pKa = 10.32LVSNLYY330 pKa = 10.55GSWAPAIMTCSPKK343 pKa = 10.65GNNGLTVEE351 pKa = 4.73FDD353 pKa = 3.32LSKK356 pKa = 11.64AEE358 pKa = 4.16VTVLTSNILKK368 pKa = 10.15VKK370 pKa = 10.15NISVDD375 pKa = 3.4GTNDD379 pKa = 3.67SILDD383 pKa = 3.63VNASPFIGKK392 pKa = 10.04GYY394 pKa = 10.15IIGGTNIEE402 pKa = 4.44FKK404 pKa = 10.52GYY406 pKa = 9.31PYY408 pKa = 10.85VIGGDD413 pKa = 3.94LNFSPDD419 pKa = 3.19TDD421 pKa = 2.87SGYY424 pKa = 8.62TVKK427 pKa = 9.85GTASVGGTKK436 pKa = 8.96IDD438 pKa = 3.8RR439 pKa = 11.84PPLKK443 pKa = 10.03ATIGRR448 pKa = 11.84CRR450 pKa = 11.84SMGNIGMNVGEE461 pKa = 4.69IKK463 pKa = 10.57SHH465 pKa = 5.75IAGYY469 pKa = 7.1SHH471 pKa = 7.56KK472 pKa = 10.89GNLWTFLTKK481 pKa = 10.65LGVRR485 pKa = 11.84SSTNAAPINLIQTRR499 pKa = 11.84SIGTTYY505 pKa = 10.18MFYY508 pKa = 10.57LYY510 pKa = 10.56KK511 pKa = 10.63QIFTDD516 pKa = 3.48NGNLSVEE523 pKa = 4.56AEE525 pKa = 4.28HH526 pKa = 6.99NMTTGVIVNLKK537 pKa = 8.58PTIHH541 pKa = 6.18SCKK544 pKa = 10.28SVNN547 pKa = 3.24
Molecular weight: 60.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
810 |
263 |
547 |
405.0 |
45.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.667 ± 0.322 | 1.358 ± 0.09 |
4.938 ± 0.213 | 3.58 ± 1.173 |
2.84 ± 0.322 | 6.914 ± 0.879 |
2.346 ± 0.175 | 6.667 ± 0.308 |
7.284 ± 0.453 | 5.802 ± 0.576 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.099 ± 0.319 | 5.432 ± 0.691 |
5.185 ± 0.286 | 1.605 ± 0.374 |
6.79 ± 0.03 | 9.136 ± 0.636 |
7.407 ± 1.572 | 6.914 ± 0.172 |
2.222 ± 0.453 | 4.815 ± 0.701 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
