
Molossus molossus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 5.3
Get precalculated fractions of proteins
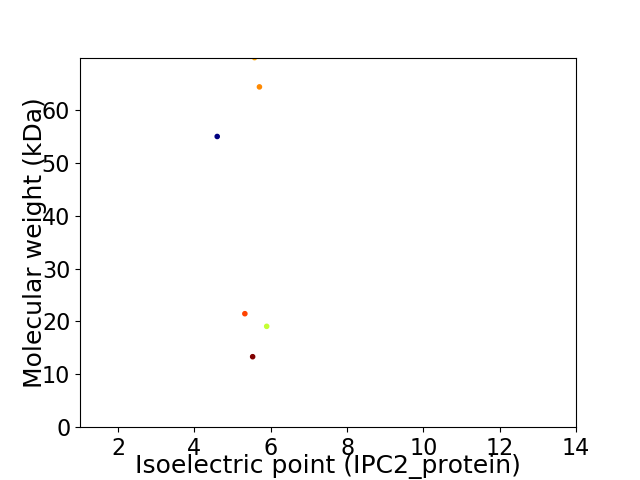
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I2MP70|A0A2I2MP70_9PAPI Protein E6 OS=Molossus molossus papillomavirus 1 OX=1959848 GN=E6 PE=3 SV=1
MM1 pKa = 7.2SRR3 pKa = 11.84AGRR6 pKa = 11.84KK7 pKa = 8.82RR8 pKa = 11.84RR9 pKa = 11.84AAAEE13 pKa = 4.52DD14 pKa = 3.69IYY16 pKa = 11.26KK17 pKa = 10.04HH18 pKa = 6.1CKK20 pKa = 9.25QFGTCPPDD28 pKa = 3.4VVNKK32 pKa = 10.3IEE34 pKa = 4.26NNTIADD40 pKa = 5.46RR41 pKa = 11.84ILKK44 pKa = 8.72WLSPFLYY51 pKa = 10.33FGGAGISSGKK61 pKa = 8.55GTGTVIPLEE70 pKa = 4.24RR71 pKa = 11.84PFGGFGSDD79 pKa = 3.41LGVAPAEE86 pKa = 4.34SGVTVGVRR94 pKa = 11.84PIPVVPAEE102 pKa = 4.39PPVGIDD108 pKa = 3.04PGGEE112 pKa = 3.94FLEE115 pKa = 4.86LEE117 pKa = 4.34PLLPVRR123 pKa = 11.84PTPEE127 pKa = 3.57VPGTVDD133 pKa = 3.45GPVDD137 pKa = 3.2TGVNVTPVVPDD148 pKa = 3.43VLEE151 pKa = 4.16VNPVNIEE158 pKa = 3.91PVEE161 pKa = 4.11PPVTVSTSLFDD172 pKa = 3.58NPTFEE177 pKa = 4.35VSITASTTTGEE188 pKa = 4.34SSAVDD193 pKa = 3.1HH194 pKa = 6.25VFVGEE199 pKa = 4.12RR200 pKa = 11.84AGGTNIGGGEE210 pKa = 4.53EE211 pKa = 4.08IPLQTFRR218 pKa = 11.84EE219 pKa = 4.4TIPDD223 pKa = 3.66DD224 pKa = 3.65EE225 pKa = 4.93FQVEE229 pKa = 4.41DD230 pKa = 3.75TGFTTSTPQEE240 pKa = 3.68GRR242 pKa = 11.84VRR244 pKa = 11.84GDD246 pKa = 2.56RR247 pKa = 11.84GYY249 pKa = 11.0LYY251 pKa = 10.67NRR253 pKa = 11.84RR254 pKa = 11.84LVQQVEE260 pKa = 4.32VPEE263 pKa = 4.33IEE265 pKa = 4.23FLEE268 pKa = 4.44EE269 pKa = 3.78PRR271 pKa = 11.84TLITFEE277 pKa = 3.95NPVYY281 pKa = 10.71NEE283 pKa = 3.79QDD285 pKa = 2.86LTMIFEE291 pKa = 4.43NEE293 pKa = 3.95LNEE296 pKa = 4.3VTRR299 pKa = 11.84TPPRR303 pKa = 11.84AAPHH307 pKa = 7.05RR308 pKa = 11.84DD309 pKa = 3.28FTDD312 pKa = 2.67IKK314 pKa = 10.73YY315 pKa = 10.52LGRR318 pKa = 11.84QLFSRR323 pKa = 11.84APKK326 pKa = 9.16SGRR329 pKa = 11.84VRR331 pKa = 11.84VSRR334 pKa = 11.84VGMKK338 pKa = 8.93GTLRR342 pKa = 11.84TRR344 pKa = 11.84SGVQIGSHH352 pKa = 3.57VHH354 pKa = 6.01YY355 pKa = 10.37YY356 pKa = 10.46QDD358 pKa = 3.86LSSILPAEE366 pKa = 4.21EE367 pKa = 4.85VEE369 pKa = 4.28LLPLGEE375 pKa = 4.22VTGTEE380 pKa = 4.17EE381 pKa = 4.39IITGEE386 pKa = 4.24GEE388 pKa = 4.33TPFVEE393 pKa = 5.22VNLNDD398 pKa = 3.85AADD401 pKa = 4.3SIGLEE406 pKa = 3.95PSNDD410 pKa = 3.14FEE412 pKa = 5.29VIEE415 pKa = 4.22NTDD418 pKa = 3.65FFGDD422 pKa = 3.52VTTTMVPDD430 pKa = 3.89PTKK433 pKa = 10.65HH434 pKa = 6.2RR435 pKa = 11.84SFYY438 pKa = 9.85PDD440 pKa = 3.01HH441 pKa = 6.98LFGFGGPNVTVDD453 pKa = 3.65YY454 pKa = 9.98PVTNIDD460 pKa = 3.1VRR462 pKa = 11.84PTVVPEE468 pKa = 3.74DD469 pKa = 4.2RR470 pKa = 11.84PWIPYY475 pKa = 10.49YY476 pKa = 10.94SFNDD480 pKa = 3.1NSTDD484 pKa = 4.29FFYY487 pKa = 10.92EE488 pKa = 3.63PCLYY492 pKa = 10.32GKK494 pKa = 9.85RR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.71RR498 pKa = 11.84CII500 pKa = 3.27
MM1 pKa = 7.2SRR3 pKa = 11.84AGRR6 pKa = 11.84KK7 pKa = 8.82RR8 pKa = 11.84RR9 pKa = 11.84AAAEE13 pKa = 4.52DD14 pKa = 3.69IYY16 pKa = 11.26KK17 pKa = 10.04HH18 pKa = 6.1CKK20 pKa = 9.25QFGTCPPDD28 pKa = 3.4VVNKK32 pKa = 10.3IEE34 pKa = 4.26NNTIADD40 pKa = 5.46RR41 pKa = 11.84ILKK44 pKa = 8.72WLSPFLYY51 pKa = 10.33FGGAGISSGKK61 pKa = 8.55GTGTVIPLEE70 pKa = 4.24RR71 pKa = 11.84PFGGFGSDD79 pKa = 3.41LGVAPAEE86 pKa = 4.34SGVTVGVRR94 pKa = 11.84PIPVVPAEE102 pKa = 4.39PPVGIDD108 pKa = 3.04PGGEE112 pKa = 3.94FLEE115 pKa = 4.86LEE117 pKa = 4.34PLLPVRR123 pKa = 11.84PTPEE127 pKa = 3.57VPGTVDD133 pKa = 3.45GPVDD137 pKa = 3.2TGVNVTPVVPDD148 pKa = 3.43VLEE151 pKa = 4.16VNPVNIEE158 pKa = 3.91PVEE161 pKa = 4.11PPVTVSTSLFDD172 pKa = 3.58NPTFEE177 pKa = 4.35VSITASTTTGEE188 pKa = 4.34SSAVDD193 pKa = 3.1HH194 pKa = 6.25VFVGEE199 pKa = 4.12RR200 pKa = 11.84AGGTNIGGGEE210 pKa = 4.53EE211 pKa = 4.08IPLQTFRR218 pKa = 11.84EE219 pKa = 4.4TIPDD223 pKa = 3.66DD224 pKa = 3.65EE225 pKa = 4.93FQVEE229 pKa = 4.41DD230 pKa = 3.75TGFTTSTPQEE240 pKa = 3.68GRR242 pKa = 11.84VRR244 pKa = 11.84GDD246 pKa = 2.56RR247 pKa = 11.84GYY249 pKa = 11.0LYY251 pKa = 10.67NRR253 pKa = 11.84RR254 pKa = 11.84LVQQVEE260 pKa = 4.32VPEE263 pKa = 4.33IEE265 pKa = 4.23FLEE268 pKa = 4.44EE269 pKa = 3.78PRR271 pKa = 11.84TLITFEE277 pKa = 3.95NPVYY281 pKa = 10.71NEE283 pKa = 3.79QDD285 pKa = 2.86LTMIFEE291 pKa = 4.43NEE293 pKa = 3.95LNEE296 pKa = 4.3VTRR299 pKa = 11.84TPPRR303 pKa = 11.84AAPHH307 pKa = 7.05RR308 pKa = 11.84DD309 pKa = 3.28FTDD312 pKa = 2.67IKK314 pKa = 10.73YY315 pKa = 10.52LGRR318 pKa = 11.84QLFSRR323 pKa = 11.84APKK326 pKa = 9.16SGRR329 pKa = 11.84VRR331 pKa = 11.84VSRR334 pKa = 11.84VGMKK338 pKa = 8.93GTLRR342 pKa = 11.84TRR344 pKa = 11.84SGVQIGSHH352 pKa = 3.57VHH354 pKa = 6.01YY355 pKa = 10.37YY356 pKa = 10.46QDD358 pKa = 3.86LSSILPAEE366 pKa = 4.21EE367 pKa = 4.85VEE369 pKa = 4.28LLPLGEE375 pKa = 4.22VTGTEE380 pKa = 4.17EE381 pKa = 4.39IITGEE386 pKa = 4.24GEE388 pKa = 4.33TPFVEE393 pKa = 5.22VNLNDD398 pKa = 3.85AADD401 pKa = 4.3SIGLEE406 pKa = 3.95PSNDD410 pKa = 3.14FEE412 pKa = 5.29VIEE415 pKa = 4.22NTDD418 pKa = 3.65FFGDD422 pKa = 3.52VTTTMVPDD430 pKa = 3.89PTKK433 pKa = 10.65HH434 pKa = 6.2RR435 pKa = 11.84SFYY438 pKa = 9.85PDD440 pKa = 3.01HH441 pKa = 6.98LFGFGGPNVTVDD453 pKa = 3.65YY454 pKa = 9.98PVTNIDD460 pKa = 3.1VRR462 pKa = 11.84PTVVPEE468 pKa = 3.74DD469 pKa = 4.2RR470 pKa = 11.84PWIPYY475 pKa = 10.49YY476 pKa = 10.94SFNDD480 pKa = 3.1NSTDD484 pKa = 4.29FFYY487 pKa = 10.92EE488 pKa = 3.63PCLYY492 pKa = 10.32GKK494 pKa = 9.85RR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.71RR498 pKa = 11.84CII500 pKa = 3.27
Molecular weight: 54.96 kDa
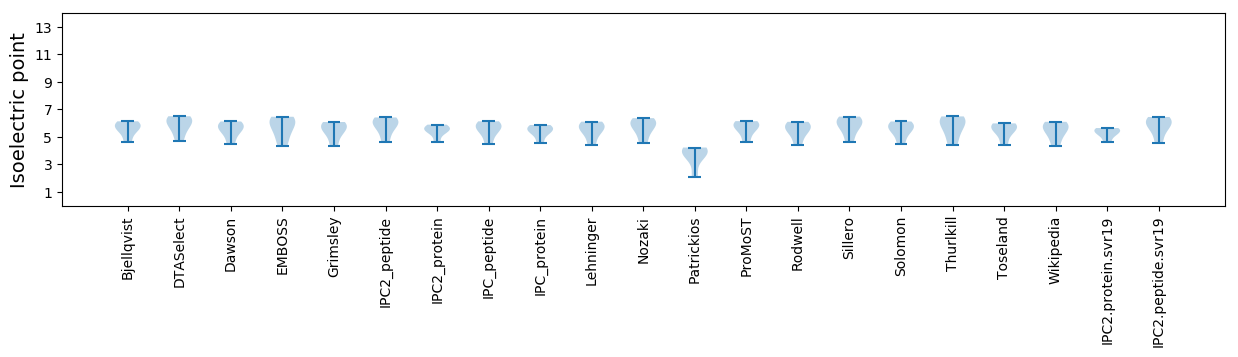
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I2MP74|A0A2I2MP74_9PAPI Major capsid protein L1 OS=Molossus molossus papillomavirus 1 OX=1959848 GN=L1 PE=3 SV=1
MM1 pKa = 7.63LFLKK5 pKa = 9.69TDD7 pKa = 3.66PGFLTTVLMTILLIFSMNLAYY28 pKa = 9.42MVKK31 pKa = 10.59GEE33 pKa = 3.97NAVFNLLQMAFWRR46 pKa = 11.84PAQNKK51 pKa = 8.99FYY53 pKa = 10.53IPPTPITRR61 pKa = 11.84AISTDD66 pKa = 3.44EE67 pKa = 4.03YY68 pKa = 9.9VHH70 pKa = 5.94RR71 pKa = 11.84TNVFYY76 pKa = 10.52HH77 pKa = 6.69ASSEE81 pKa = 4.02RR82 pKa = 11.84LLMVGHH88 pKa = 7.64PYY90 pKa = 10.49FEE92 pKa = 4.71IPSEE96 pKa = 4.13NDD98 pKa = 3.05DD99 pKa = 3.73APAPKK104 pKa = 10.08VPKK107 pKa = 10.24VSPNQYY113 pKa = 9.11RR114 pKa = 11.84VFRR117 pKa = 11.84IKK119 pKa = 10.74LPNPNEE125 pKa = 3.85FVFPDD130 pKa = 3.44KK131 pKa = 10.88NAYY134 pKa = 10.04NPDD137 pKa = 3.47TEE139 pKa = 4.21RR140 pKa = 11.84LVWACRR146 pKa = 11.84GLEE149 pKa = 3.96VGRR152 pKa = 11.84GGPLGVAVTGSPFFNKK168 pKa = 10.02LHH170 pKa = 6.85DD171 pKa = 3.98VEE173 pKa = 5.49NPLNGAYY180 pKa = 9.91EE181 pKa = 4.27PEE183 pKa = 3.99DD184 pKa = 3.44TKK186 pKa = 11.45NEE188 pKa = 3.97RR189 pKa = 11.84RR190 pKa = 11.84NMAFDD195 pKa = 4.43CKK197 pKa = 8.8QTQLFIVGSNPPLGEE212 pKa = 3.82YY213 pKa = 6.68WTAAKK218 pKa = 10.15PCGEE222 pKa = 4.75LKK224 pKa = 10.95AGDD227 pKa = 4.35CPPLEE232 pKa = 4.76LLDD235 pKa = 4.46KK236 pKa = 10.91VIEE239 pKa = 4.82DD240 pKa = 4.08GQMCDD245 pKa = 3.13IGFGNIDD252 pKa = 3.99AKK254 pKa = 10.7RR255 pKa = 11.84LQPNKK260 pKa = 10.51SEE262 pKa = 4.6LPLDD266 pKa = 4.43SIDD269 pKa = 5.85EE270 pKa = 4.61IICYY274 pKa = 9.45PDD276 pKa = 3.31YY277 pKa = 11.53HH278 pKa = 8.4KK279 pKa = 8.92MTRR282 pKa = 11.84DD283 pKa = 3.14LFGNYY288 pKa = 7.2LWFFARR294 pKa = 11.84RR295 pKa = 11.84EE296 pKa = 3.9QMYY299 pKa = 10.35ARR301 pKa = 11.84HH302 pKa = 5.81FMDD305 pKa = 3.99RR306 pKa = 11.84AGEE309 pKa = 4.09MKK311 pKa = 9.63EE312 pKa = 4.21TIPEE316 pKa = 4.13SLYY319 pKa = 10.66LAPANEE325 pKa = 4.44DD326 pKa = 3.5QNTGPGQLNSLAPYY340 pKa = 9.92SYY342 pKa = 9.34TATPSGSLVSTDD354 pKa = 2.92TQFFNRR360 pKa = 11.84PYY362 pKa = 10.0WLNKK366 pKa = 9.6AQGLNNGICWNNQLFVTVMDD386 pKa = 3.82NTRR389 pKa = 11.84GTNFTINVNSAEE401 pKa = 4.27GSLTEE406 pKa = 4.42YY407 pKa = 10.89DD408 pKa = 3.0ATKK411 pKa = 9.49INEE414 pKa = 4.16YY415 pKa = 9.95LRR417 pKa = 11.84HH418 pKa = 5.99VEE420 pKa = 3.96EE421 pKa = 5.21FEE423 pKa = 5.0LSFIFQLCRR432 pKa = 11.84VSLSAEE438 pKa = 3.57ILSYY442 pKa = 10.58VHH444 pKa = 6.54TMDD447 pKa = 4.8STIIDD452 pKa = 3.49NWDD455 pKa = 3.55LNIASPPNDD464 pKa = 3.8SLHH467 pKa = 6.89EE468 pKa = 4.04IYY470 pKa = 10.34RR471 pKa = 11.84WIEE474 pKa = 3.93SQATKK479 pKa = 10.74CPDD482 pKa = 3.14AVKK485 pKa = 10.19PPEE488 pKa = 5.71DD489 pKa = 3.66KK490 pKa = 11.22DD491 pKa = 3.12PWKK494 pKa = 10.45DD495 pKa = 3.51YY496 pKa = 11.23KK497 pKa = 10.66FWNVDD502 pKa = 3.02LTEE505 pKa = 4.54RR506 pKa = 11.84FSEE509 pKa = 5.2QLDD512 pKa = 3.6QYY514 pKa = 11.23NLGRR518 pKa = 11.84RR519 pKa = 11.84FLFQAGLTTNQGARR533 pKa = 11.84QVVARR538 pKa = 11.84PVRR541 pKa = 11.84TVRR544 pKa = 11.84TVRR547 pKa = 11.84TARR550 pKa = 11.84AVKK553 pKa = 10.22RR554 pKa = 11.84KK555 pKa = 9.35RR556 pKa = 11.84SKK558 pKa = 10.23ISKK561 pKa = 9.71KK562 pKa = 10.5
MM1 pKa = 7.63LFLKK5 pKa = 9.69TDD7 pKa = 3.66PGFLTTVLMTILLIFSMNLAYY28 pKa = 9.42MVKK31 pKa = 10.59GEE33 pKa = 3.97NAVFNLLQMAFWRR46 pKa = 11.84PAQNKK51 pKa = 8.99FYY53 pKa = 10.53IPPTPITRR61 pKa = 11.84AISTDD66 pKa = 3.44EE67 pKa = 4.03YY68 pKa = 9.9VHH70 pKa = 5.94RR71 pKa = 11.84TNVFYY76 pKa = 10.52HH77 pKa = 6.69ASSEE81 pKa = 4.02RR82 pKa = 11.84LLMVGHH88 pKa = 7.64PYY90 pKa = 10.49FEE92 pKa = 4.71IPSEE96 pKa = 4.13NDD98 pKa = 3.05DD99 pKa = 3.73APAPKK104 pKa = 10.08VPKK107 pKa = 10.24VSPNQYY113 pKa = 9.11RR114 pKa = 11.84VFRR117 pKa = 11.84IKK119 pKa = 10.74LPNPNEE125 pKa = 3.85FVFPDD130 pKa = 3.44KK131 pKa = 10.88NAYY134 pKa = 10.04NPDD137 pKa = 3.47TEE139 pKa = 4.21RR140 pKa = 11.84LVWACRR146 pKa = 11.84GLEE149 pKa = 3.96VGRR152 pKa = 11.84GGPLGVAVTGSPFFNKK168 pKa = 10.02LHH170 pKa = 6.85DD171 pKa = 3.98VEE173 pKa = 5.49NPLNGAYY180 pKa = 9.91EE181 pKa = 4.27PEE183 pKa = 3.99DD184 pKa = 3.44TKK186 pKa = 11.45NEE188 pKa = 3.97RR189 pKa = 11.84RR190 pKa = 11.84NMAFDD195 pKa = 4.43CKK197 pKa = 8.8QTQLFIVGSNPPLGEE212 pKa = 3.82YY213 pKa = 6.68WTAAKK218 pKa = 10.15PCGEE222 pKa = 4.75LKK224 pKa = 10.95AGDD227 pKa = 4.35CPPLEE232 pKa = 4.76LLDD235 pKa = 4.46KK236 pKa = 10.91VIEE239 pKa = 4.82DD240 pKa = 4.08GQMCDD245 pKa = 3.13IGFGNIDD252 pKa = 3.99AKK254 pKa = 10.7RR255 pKa = 11.84LQPNKK260 pKa = 10.51SEE262 pKa = 4.6LPLDD266 pKa = 4.43SIDD269 pKa = 5.85EE270 pKa = 4.61IICYY274 pKa = 9.45PDD276 pKa = 3.31YY277 pKa = 11.53HH278 pKa = 8.4KK279 pKa = 8.92MTRR282 pKa = 11.84DD283 pKa = 3.14LFGNYY288 pKa = 7.2LWFFARR294 pKa = 11.84RR295 pKa = 11.84EE296 pKa = 3.9QMYY299 pKa = 10.35ARR301 pKa = 11.84HH302 pKa = 5.81FMDD305 pKa = 3.99RR306 pKa = 11.84AGEE309 pKa = 4.09MKK311 pKa = 9.63EE312 pKa = 4.21TIPEE316 pKa = 4.13SLYY319 pKa = 10.66LAPANEE325 pKa = 4.44DD326 pKa = 3.5QNTGPGQLNSLAPYY340 pKa = 9.92SYY342 pKa = 9.34TATPSGSLVSTDD354 pKa = 2.92TQFFNRR360 pKa = 11.84PYY362 pKa = 10.0WLNKK366 pKa = 9.6AQGLNNGICWNNQLFVTVMDD386 pKa = 3.82NTRR389 pKa = 11.84GTNFTINVNSAEE401 pKa = 4.27GSLTEE406 pKa = 4.42YY407 pKa = 10.89DD408 pKa = 3.0ATKK411 pKa = 9.49INEE414 pKa = 4.16YY415 pKa = 9.95LRR417 pKa = 11.84HH418 pKa = 5.99VEE420 pKa = 3.96EE421 pKa = 5.21FEE423 pKa = 5.0LSFIFQLCRR432 pKa = 11.84VSLSAEE438 pKa = 3.57ILSYY442 pKa = 10.58VHH444 pKa = 6.54TMDD447 pKa = 4.8STIIDD452 pKa = 3.49NWDD455 pKa = 3.55LNIASPPNDD464 pKa = 3.8SLHH467 pKa = 6.89EE468 pKa = 4.04IYY470 pKa = 10.34RR471 pKa = 11.84WIEE474 pKa = 3.93SQATKK479 pKa = 10.74CPDD482 pKa = 3.14AVKK485 pKa = 10.19PPEE488 pKa = 5.71DD489 pKa = 3.66KK490 pKa = 11.22DD491 pKa = 3.12PWKK494 pKa = 10.45DD495 pKa = 3.51YY496 pKa = 11.23KK497 pKa = 10.66FWNVDD502 pKa = 3.02LTEE505 pKa = 4.54RR506 pKa = 11.84FSEE509 pKa = 5.2QLDD512 pKa = 3.6QYY514 pKa = 11.23NLGRR518 pKa = 11.84RR519 pKa = 11.84FLFQAGLTTNQGARR533 pKa = 11.84QVVARR538 pKa = 11.84PVRR541 pKa = 11.84TVRR544 pKa = 11.84TVRR547 pKa = 11.84TARR550 pKa = 11.84AVKK553 pKa = 10.22RR554 pKa = 11.84KK555 pKa = 9.35RR556 pKa = 11.84SKK558 pKa = 10.23ISKK561 pKa = 9.71KK562 pKa = 10.5
Molecular weight: 64.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2145 |
119 |
613 |
357.5 |
40.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.061 ± 0.624 | 2.564 ± 0.813 |
5.967 ± 0.486 | 7.413 ± 0.697 |
5.548 ± 0.351 | 6.014 ± 0.84 |
1.772 ± 0.179 | 4.895 ± 0.146 |
5.128 ± 0.761 | 8.998 ± 0.933 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.865 ± 0.295 | 4.476 ± 0.849 |
6.34 ± 1.036 | 3.823 ± 0.542 |
5.641 ± 0.426 | 6.34 ± 0.795 |
5.967 ± 0.768 | 6.247 ± 1.291 |
1.305 ± 0.311 | 3.636 ± 0.231 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
