
Croceicoccus marinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Croceicoccus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
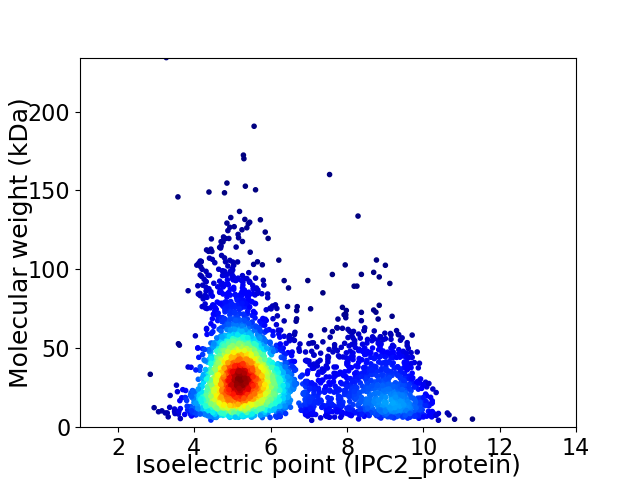
Virtual 2D-PAGE plot for 3600 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
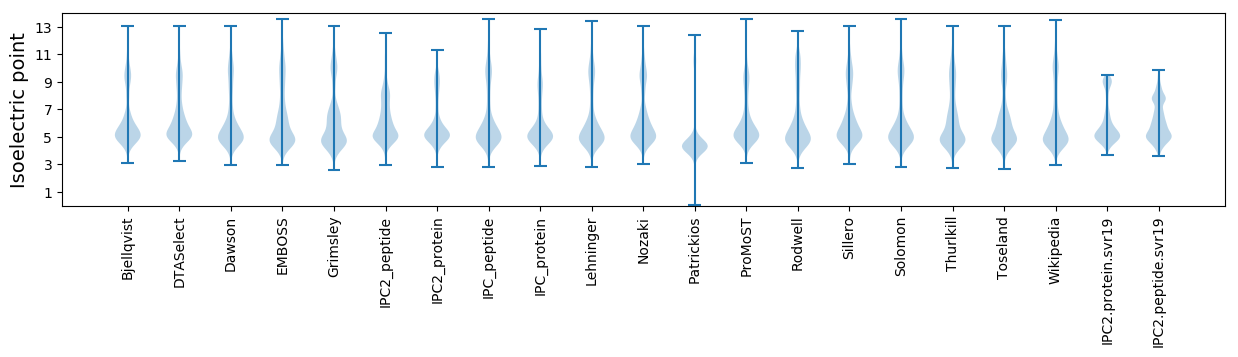
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z1FEV2|A0A1Z1FEV2_9SPHN Tyr recombinase domain-containing protein OS=Croceicoccus marinus OX=450378 GN=A9D14_05040 PE=4 SV=1
MM1 pKa = 7.48LALAGCGGDD10 pKa = 3.63QIVSPGTGGDD20 pKa = 3.63IIIEE24 pKa = 4.29NPAPAPAPTPSPTPTPTPMVTPAAGCPTINSSGGLTDD61 pKa = 4.4AGTITGDD68 pKa = 2.94TGEE71 pKa = 4.36YY72 pKa = 9.44RR73 pKa = 11.84VCTLPASFDD82 pKa = 3.57EE83 pKa = 4.75SDD85 pKa = 3.34NLPYY89 pKa = 10.0IEE91 pKa = 4.77GLLYY95 pKa = 10.59SINGRR100 pKa = 11.84VDD102 pKa = 2.73VGTDD106 pKa = 3.31GGAVPSDD113 pKa = 3.6ADD115 pKa = 3.53SDD117 pKa = 4.07VTLSIEE123 pKa = 4.36PGAILYY129 pKa = 8.11GATGRR134 pKa = 11.84SYY136 pKa = 11.35LVVNRR141 pKa = 11.84GNQIDD146 pKa = 4.09AVGEE150 pKa = 3.73PDD152 pKa = 3.42MPIVFTSRR160 pKa = 11.84DD161 pKa = 3.59NVLGLSTDD169 pKa = 3.28GSDD172 pKa = 3.72AQWGGVVLLGRR183 pKa = 11.84APVSDD188 pKa = 4.3CRR190 pKa = 11.84DD191 pKa = 3.44GVPNKK196 pKa = 10.24ANQASCEE203 pKa = 3.94QALEE207 pKa = 4.32GAAVEE212 pKa = 4.4TLFGGNTPGDD222 pKa = 3.6SSGTMQYY229 pKa = 10.61FQIRR233 pKa = 11.84YY234 pKa = 9.88SGFTLEE240 pKa = 5.43GGSEE244 pKa = 4.14LQSLTTGGIGSGTTIDD260 pKa = 3.05HH261 pKa = 5.9WMSFNSSDD269 pKa = 4.62DD270 pKa = 3.31GTEE273 pKa = 4.01FFGGRR278 pKa = 11.84VNMKK282 pKa = 9.47NVVVVGASDD291 pKa = 4.34DD292 pKa = 3.98SLDD295 pKa = 3.57VDD297 pKa = 4.44TGAQANLQYY306 pKa = 11.14VIVAQRR312 pKa = 11.84ANTGDD317 pKa = 3.94SIIEE321 pKa = 4.05LDD323 pKa = 5.24SPDD326 pKa = 3.72EE327 pKa = 4.75DD328 pKa = 4.47YY329 pKa = 7.86TTGALPRR336 pKa = 11.84TNLQIANATFLSRR349 pKa = 11.84STGSSQALRR358 pKa = 11.84LRR360 pKa = 11.84GGAQFSLVNSVIDD373 pKa = 3.49IEE375 pKa = 4.36GDD377 pKa = 3.61RR378 pKa = 11.84TCVRR382 pKa = 11.84LDD384 pKa = 3.37EE385 pKa = 4.57TVTLAQDD392 pKa = 3.45PLFASVVADD401 pKa = 4.27CGTTQPFRR409 pKa = 11.84GSSGVTDD416 pKa = 4.03AQVAAAFNDD425 pKa = 3.76GDD427 pKa = 3.92DD428 pKa = 3.68TDD430 pKa = 3.91AAFAITLTGSFINGTGEE447 pKa = 4.1TGVAAYY453 pKa = 10.44NPTALSSYY461 pKa = 10.76FDD463 pKa = 3.54DD464 pKa = 3.48TDD466 pKa = 4.17YY467 pKa = 11.35IGAVADD473 pKa = 4.4QTDD476 pKa = 3.03TWYY479 pKa = 10.37EE480 pKa = 3.69GWTCDD485 pKa = 4.7SATLTFGNNTGPCTEE500 pKa = 4.49LPVYY504 pKa = 10.5SS505 pKa = 4.15
MM1 pKa = 7.48LALAGCGGDD10 pKa = 3.63QIVSPGTGGDD20 pKa = 3.63IIIEE24 pKa = 4.29NPAPAPAPTPSPTPTPTPMVTPAAGCPTINSSGGLTDD61 pKa = 4.4AGTITGDD68 pKa = 2.94TGEE71 pKa = 4.36YY72 pKa = 9.44RR73 pKa = 11.84VCTLPASFDD82 pKa = 3.57EE83 pKa = 4.75SDD85 pKa = 3.34NLPYY89 pKa = 10.0IEE91 pKa = 4.77GLLYY95 pKa = 10.59SINGRR100 pKa = 11.84VDD102 pKa = 2.73VGTDD106 pKa = 3.31GGAVPSDD113 pKa = 3.6ADD115 pKa = 3.53SDD117 pKa = 4.07VTLSIEE123 pKa = 4.36PGAILYY129 pKa = 8.11GATGRR134 pKa = 11.84SYY136 pKa = 11.35LVVNRR141 pKa = 11.84GNQIDD146 pKa = 4.09AVGEE150 pKa = 3.73PDD152 pKa = 3.42MPIVFTSRR160 pKa = 11.84DD161 pKa = 3.59NVLGLSTDD169 pKa = 3.28GSDD172 pKa = 3.72AQWGGVVLLGRR183 pKa = 11.84APVSDD188 pKa = 4.3CRR190 pKa = 11.84DD191 pKa = 3.44GVPNKK196 pKa = 10.24ANQASCEE203 pKa = 3.94QALEE207 pKa = 4.32GAAVEE212 pKa = 4.4TLFGGNTPGDD222 pKa = 3.6SSGTMQYY229 pKa = 10.61FQIRR233 pKa = 11.84YY234 pKa = 9.88SGFTLEE240 pKa = 5.43GGSEE244 pKa = 4.14LQSLTTGGIGSGTTIDD260 pKa = 3.05HH261 pKa = 5.9WMSFNSSDD269 pKa = 4.62DD270 pKa = 3.31GTEE273 pKa = 4.01FFGGRR278 pKa = 11.84VNMKK282 pKa = 9.47NVVVVGASDD291 pKa = 4.34DD292 pKa = 3.98SLDD295 pKa = 3.57VDD297 pKa = 4.44TGAQANLQYY306 pKa = 11.14VIVAQRR312 pKa = 11.84ANTGDD317 pKa = 3.94SIIEE321 pKa = 4.05LDD323 pKa = 5.24SPDD326 pKa = 3.72EE327 pKa = 4.75DD328 pKa = 4.47YY329 pKa = 7.86TTGALPRR336 pKa = 11.84TNLQIANATFLSRR349 pKa = 11.84STGSSQALRR358 pKa = 11.84LRR360 pKa = 11.84GGAQFSLVNSVIDD373 pKa = 3.49IEE375 pKa = 4.36GDD377 pKa = 3.61RR378 pKa = 11.84TCVRR382 pKa = 11.84LDD384 pKa = 3.37EE385 pKa = 4.57TVTLAQDD392 pKa = 3.45PLFASVVADD401 pKa = 4.27CGTTQPFRR409 pKa = 11.84GSSGVTDD416 pKa = 4.03AQVAAAFNDD425 pKa = 3.76GDD427 pKa = 3.92DD428 pKa = 3.68TDD430 pKa = 3.91AAFAITLTGSFINGTGEE447 pKa = 4.1TGVAAYY453 pKa = 10.44NPTALSSYY461 pKa = 10.76FDD463 pKa = 3.54DD464 pKa = 3.48TDD466 pKa = 4.17YY467 pKa = 11.35IGAVADD473 pKa = 4.4QTDD476 pKa = 3.03TWYY479 pKa = 10.37EE480 pKa = 3.69GWTCDD485 pKa = 4.7SATLTFGNNTGPCTEE500 pKa = 4.49LPVYY504 pKa = 10.5SS505 pKa = 4.15
Molecular weight: 51.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z1FDW3|A0A1Z1FDW3_9SPHN Transcription termination factor Rho OS=Croceicoccus marinus OX=450378 GN=rho PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84TRR21 pKa = 11.84SATPGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84TRR21 pKa = 11.84SATPGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1160639 |
38 |
2329 |
322.4 |
34.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.762 ± 0.06 | 0.841 ± 0.014 |
6.374 ± 0.042 | 6.05 ± 0.04 |
3.596 ± 0.027 | 8.888 ± 0.038 |
2.036 ± 0.022 | 5.02 ± 0.026 |
2.847 ± 0.032 | 9.716 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.654 ± 0.023 | 2.535 ± 0.018 |
5.254 ± 0.027 | 3.201 ± 0.023 |
7.379 ± 0.038 | 5.318 ± 0.027 |
4.971 ± 0.029 | 6.92 ± 0.029 |
1.437 ± 0.018 | 2.201 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
