
Viral hemorrhagic septicemia virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Gammarhabdovirinae; Novirhabdovirus; Piscine novirhabdovirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
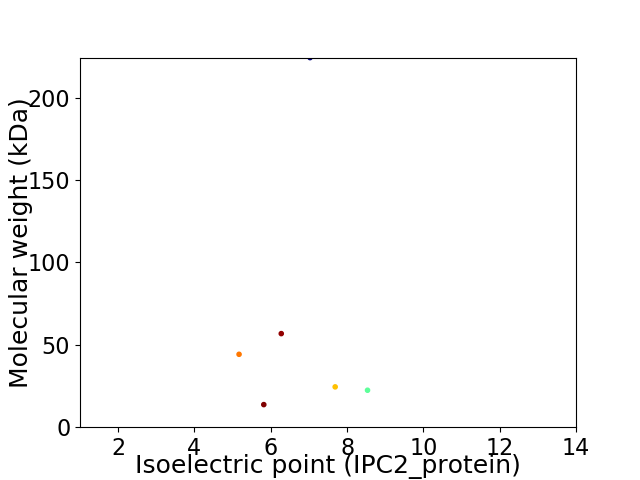
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
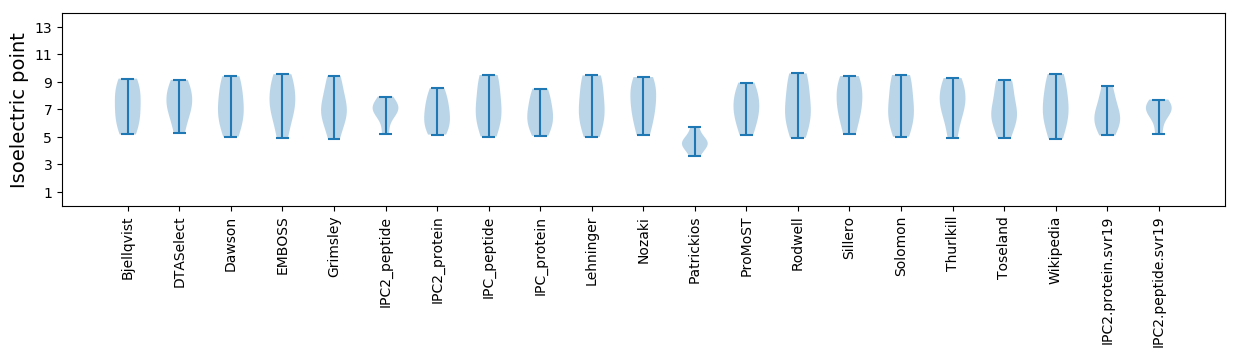
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S4VAX6|S4VAX6_9RHAB GDP polyribonucleotidyltransferase OS=Viral hemorrhagic septicemia virus OX=11287 GN=L PE=4 SV=1
MM1 pKa = 7.61EE2 pKa = 5.39GGLRR6 pKa = 11.84AAFSGLNDD14 pKa = 3.32VRR16 pKa = 11.84IDD18 pKa = 3.5PTGGEE23 pKa = 4.17GRR25 pKa = 11.84VLVPGDD31 pKa = 3.53VEE33 pKa = 4.97LIVYY37 pKa = 9.72VGGFGEE43 pKa = 4.44EE44 pKa = 3.92DD45 pKa = 3.19RR46 pKa = 11.84KK47 pKa = 11.18VIVDD51 pKa = 3.84ALSALGGPQTVQALSVLLSYY71 pKa = 10.6VLQGNTQEE79 pKa = 5.28DD80 pKa = 5.29LEE82 pKa = 4.86TKK84 pKa = 10.55CKK86 pKa = 10.67VLTDD90 pKa = 3.37MGFKK94 pKa = 8.85VTQAVRR100 pKa = 11.84ATSIEE105 pKa = 4.05AGIMMPMRR113 pKa = 11.84EE114 pKa = 4.11LALTVNDD121 pKa = 4.5DD122 pKa = 3.6NLMEE126 pKa = 4.42IVKK129 pKa = 8.57GTLMTCSLLTKK140 pKa = 10.37YY141 pKa = 10.81SVDD144 pKa = 3.72KK145 pKa = 9.66MIKK148 pKa = 10.37YY149 pKa = 7.92ITKK152 pKa = 10.55KK153 pKa = 10.45LGEE156 pKa = 4.24LADD159 pKa = 3.93TQGVGEE165 pKa = 4.55LQHH168 pKa = 5.73FTADD172 pKa = 3.03KK173 pKa = 10.57AAIRR177 pKa = 11.84KK178 pKa = 8.49LAGCVRR184 pKa = 11.84PGQKK188 pKa = 7.96ITKK191 pKa = 9.34ALYY194 pKa = 10.43AFILTEE200 pKa = 3.84IADD203 pKa = 3.77PTTQSRR209 pKa = 11.84ARR211 pKa = 11.84AMGALRR217 pKa = 11.84LNGTGMTMIGLFTQAANNLGIAPAKK242 pKa = 10.36LLEE245 pKa = 4.44DD246 pKa = 3.75LCMEE250 pKa = 4.52SLVEE254 pKa = 3.87SARR257 pKa = 11.84RR258 pKa = 11.84IIQLMRR264 pKa = 11.84QVSEE268 pKa = 3.86AKK270 pKa = 10.34SIQEE274 pKa = 4.01RR275 pKa = 11.84YY276 pKa = 10.18AIMMSRR282 pKa = 11.84MLGEE286 pKa = 4.49SYY288 pKa = 10.77YY289 pKa = 11.0KK290 pKa = 10.58SYY292 pKa = 10.83GLNDD296 pKa = 3.4NSKK299 pKa = 9.96ISYY302 pKa = 9.71ILSQISGKK310 pKa = 10.03YY311 pKa = 9.9AVDD314 pKa = 3.4SLEE317 pKa = 3.87GLEE320 pKa = 4.64GIKK323 pKa = 9.22VTEE326 pKa = 4.2KK327 pKa = 10.21FRR329 pKa = 11.84EE330 pKa = 4.06FAEE333 pKa = 4.36LVAEE337 pKa = 4.19VLVDD341 pKa = 3.29KK342 pKa = 10.41YY343 pKa = 11.34EE344 pKa = 4.77RR345 pKa = 11.84IGEE348 pKa = 4.24DD349 pKa = 3.07STEE352 pKa = 3.94VSDD355 pKa = 5.0VIKK358 pKa = 10.46EE359 pKa = 3.92AARR362 pKa = 11.84QHH364 pKa = 5.75ARR366 pKa = 11.84RR367 pKa = 11.84TSAKK371 pKa = 9.81PEE373 pKa = 3.73PKK375 pKa = 10.08ARR377 pKa = 11.84NFRR380 pKa = 11.84SSTGRR385 pKa = 11.84GKK387 pKa = 9.68EE388 pKa = 3.93QEE390 pKa = 4.28TGEE393 pKa = 4.59SDD395 pKa = 4.29DD396 pKa = 5.12DD397 pKa = 5.31DD398 pKa = 4.73YY399 pKa = 12.03PEE401 pKa = 6.09DD402 pKa = 3.8SDD404 pKa = 4.34
MM1 pKa = 7.61EE2 pKa = 5.39GGLRR6 pKa = 11.84AAFSGLNDD14 pKa = 3.32VRR16 pKa = 11.84IDD18 pKa = 3.5PTGGEE23 pKa = 4.17GRR25 pKa = 11.84VLVPGDD31 pKa = 3.53VEE33 pKa = 4.97LIVYY37 pKa = 9.72VGGFGEE43 pKa = 4.44EE44 pKa = 3.92DD45 pKa = 3.19RR46 pKa = 11.84KK47 pKa = 11.18VIVDD51 pKa = 3.84ALSALGGPQTVQALSVLLSYY71 pKa = 10.6VLQGNTQEE79 pKa = 5.28DD80 pKa = 5.29LEE82 pKa = 4.86TKK84 pKa = 10.55CKK86 pKa = 10.67VLTDD90 pKa = 3.37MGFKK94 pKa = 8.85VTQAVRR100 pKa = 11.84ATSIEE105 pKa = 4.05AGIMMPMRR113 pKa = 11.84EE114 pKa = 4.11LALTVNDD121 pKa = 4.5DD122 pKa = 3.6NLMEE126 pKa = 4.42IVKK129 pKa = 8.57GTLMTCSLLTKK140 pKa = 10.37YY141 pKa = 10.81SVDD144 pKa = 3.72KK145 pKa = 9.66MIKK148 pKa = 10.37YY149 pKa = 7.92ITKK152 pKa = 10.55KK153 pKa = 10.45LGEE156 pKa = 4.24LADD159 pKa = 3.93TQGVGEE165 pKa = 4.55LQHH168 pKa = 5.73FTADD172 pKa = 3.03KK173 pKa = 10.57AAIRR177 pKa = 11.84KK178 pKa = 8.49LAGCVRR184 pKa = 11.84PGQKK188 pKa = 7.96ITKK191 pKa = 9.34ALYY194 pKa = 10.43AFILTEE200 pKa = 3.84IADD203 pKa = 3.77PTTQSRR209 pKa = 11.84ARR211 pKa = 11.84AMGALRR217 pKa = 11.84LNGTGMTMIGLFTQAANNLGIAPAKK242 pKa = 10.36LLEE245 pKa = 4.44DD246 pKa = 3.75LCMEE250 pKa = 4.52SLVEE254 pKa = 3.87SARR257 pKa = 11.84RR258 pKa = 11.84IIQLMRR264 pKa = 11.84QVSEE268 pKa = 3.86AKK270 pKa = 10.34SIQEE274 pKa = 4.01RR275 pKa = 11.84YY276 pKa = 10.18AIMMSRR282 pKa = 11.84MLGEE286 pKa = 4.49SYY288 pKa = 10.77YY289 pKa = 11.0KK290 pKa = 10.58SYY292 pKa = 10.83GLNDD296 pKa = 3.4NSKK299 pKa = 9.96ISYY302 pKa = 9.71ILSQISGKK310 pKa = 10.03YY311 pKa = 9.9AVDD314 pKa = 3.4SLEE317 pKa = 3.87GLEE320 pKa = 4.64GIKK323 pKa = 9.22VTEE326 pKa = 4.2KK327 pKa = 10.21FRR329 pKa = 11.84EE330 pKa = 4.06FAEE333 pKa = 4.36LVAEE337 pKa = 4.19VLVDD341 pKa = 3.29KK342 pKa = 10.41YY343 pKa = 11.34EE344 pKa = 4.77RR345 pKa = 11.84IGEE348 pKa = 4.24DD349 pKa = 3.07STEE352 pKa = 3.94VSDD355 pKa = 5.0VIKK358 pKa = 10.46EE359 pKa = 3.92AARR362 pKa = 11.84QHH364 pKa = 5.75ARR366 pKa = 11.84RR367 pKa = 11.84TSAKK371 pKa = 9.81PEE373 pKa = 3.73PKK375 pKa = 10.08ARR377 pKa = 11.84NFRR380 pKa = 11.84SSTGRR385 pKa = 11.84GKK387 pKa = 9.68EE388 pKa = 3.93QEE390 pKa = 4.28TGEE393 pKa = 4.59SDD395 pKa = 4.29DD396 pKa = 5.12DD397 pKa = 5.31DD398 pKa = 4.73YY399 pKa = 12.03PEE401 pKa = 6.09DD402 pKa = 3.8SDD404 pKa = 4.34
Molecular weight: 44.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q08489|Q08489_9RHAB Non-virion protein OS=Viral hemorrhagic septicemia virus OX=11287 GN=NV PE=4 SV=1
MM1 pKa = 7.42TLFKK5 pKa = 10.38RR6 pKa = 11.84KK7 pKa = 8.44RR8 pKa = 11.84TILVPPPHH16 pKa = 6.44LTSNDD21 pKa = 3.32EE22 pKa = 4.34DD23 pKa = 3.71RR24 pKa = 11.84VSTILTEE31 pKa = 4.17GTLTITGPPPGNQVDD46 pKa = 4.38KK47 pKa = 11.47VCMAMKK53 pKa = 10.22LARR56 pKa = 11.84AILCEE61 pKa = 4.17DD62 pKa = 3.14QHH64 pKa = 6.64PAFNPLVHH72 pKa = 6.97LFQSAMIFGEE82 pKa = 4.23TSEE85 pKa = 5.33KK86 pKa = 10.04IDD88 pKa = 3.42FGTRR92 pKa = 11.84SKK94 pKa = 9.83TLITSFKK101 pKa = 9.8IAEE104 pKa = 4.22AKK106 pKa = 10.35AIYY109 pKa = 10.03LDD111 pKa = 3.81SSPVRR116 pKa = 11.84SRR118 pKa = 11.84IEE120 pKa = 3.74AKK122 pKa = 10.43KK123 pKa = 7.99YY124 pKa = 4.56TTPIRR129 pKa = 11.84HH130 pKa = 6.09GSVTYY135 pKa = 9.47YY136 pKa = 11.16GPFIFADD143 pKa = 3.49DD144 pKa = 3.77HH145 pKa = 7.49VGGKK149 pKa = 7.29GHH151 pKa = 6.61RR152 pKa = 11.84EE153 pKa = 3.6KK154 pKa = 11.12LGALCGFLQSEE165 pKa = 5.24PYY167 pKa = 10.24GQAKK171 pKa = 10.21DD172 pKa = 3.79YY173 pKa = 10.74YY174 pKa = 8.74NRR176 pKa = 11.84AVEE179 pKa = 4.22EE180 pKa = 4.47EE181 pKa = 4.0IGIPPRR187 pKa = 11.84DD188 pKa = 3.64PKK190 pKa = 10.69RR191 pKa = 11.84RR192 pKa = 11.84SGTSSVRR199 pKa = 11.84PWW201 pKa = 2.81
MM1 pKa = 7.42TLFKK5 pKa = 10.38RR6 pKa = 11.84KK7 pKa = 8.44RR8 pKa = 11.84TILVPPPHH16 pKa = 6.44LTSNDD21 pKa = 3.32EE22 pKa = 4.34DD23 pKa = 3.71RR24 pKa = 11.84VSTILTEE31 pKa = 4.17GTLTITGPPPGNQVDD46 pKa = 4.38KK47 pKa = 11.47VCMAMKK53 pKa = 10.22LARR56 pKa = 11.84AILCEE61 pKa = 4.17DD62 pKa = 3.14QHH64 pKa = 6.64PAFNPLVHH72 pKa = 6.97LFQSAMIFGEE82 pKa = 4.23TSEE85 pKa = 5.33KK86 pKa = 10.04IDD88 pKa = 3.42FGTRR92 pKa = 11.84SKK94 pKa = 9.83TLITSFKK101 pKa = 9.8IAEE104 pKa = 4.22AKK106 pKa = 10.35AIYY109 pKa = 10.03LDD111 pKa = 3.81SSPVRR116 pKa = 11.84SRR118 pKa = 11.84IEE120 pKa = 3.74AKK122 pKa = 10.43KK123 pKa = 7.99YY124 pKa = 4.56TTPIRR129 pKa = 11.84HH130 pKa = 6.09GSVTYY135 pKa = 9.47YY136 pKa = 11.16GPFIFADD143 pKa = 3.49DD144 pKa = 3.77HH145 pKa = 7.49VGGKK149 pKa = 7.29GHH151 pKa = 6.61RR152 pKa = 11.84EE153 pKa = 3.6KK154 pKa = 11.12LGALCGFLQSEE165 pKa = 5.24PYY167 pKa = 10.24GQAKK171 pKa = 10.21DD172 pKa = 3.79YY173 pKa = 10.74YY174 pKa = 8.74NRR176 pKa = 11.84AVEE179 pKa = 4.22EE180 pKa = 4.47EE181 pKa = 4.0IGIPPRR187 pKa = 11.84DD188 pKa = 3.64PKK190 pKa = 10.69RR191 pKa = 11.84RR192 pKa = 11.84SGTSSVRR199 pKa = 11.84PWW201 pKa = 2.81
Molecular weight: 22.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3440 |
122 |
1984 |
573.3 |
64.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.337 ± 0.592 | 1.483 ± 0.357 |
5.349 ± 0.29 | 6.86 ± 0.815 |
3.372 ± 0.263 | 6.279 ± 0.461 |
2.645 ± 0.527 | 6.221 ± 0.215 |
6.483 ± 0.613 | 10.814 ± 1.003 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.471 ± 0.279 | 3.052 ± 0.33 |
4.593 ± 0.752 | 3.866 ± 0.503 |
5.64 ± 0.553 | 7.645 ± 0.322 |
6.948 ± 0.572 | 5.901 ± 0.483 |
1.308 ± 0.418 | 2.733 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
